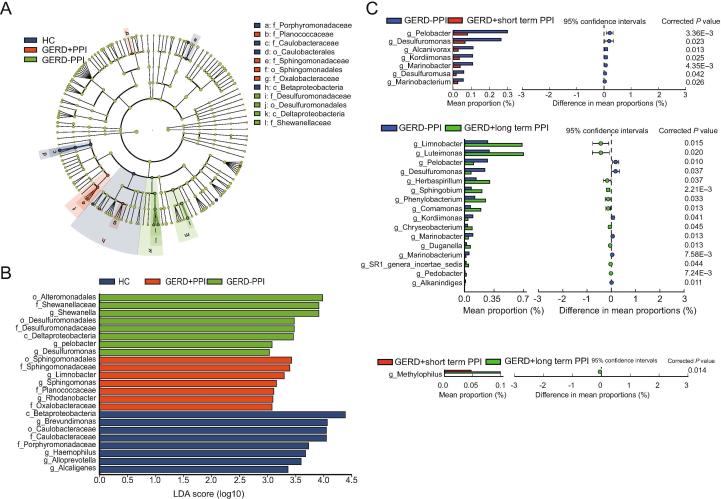
Figure 4.
Variations in the gastric mucosal microbiota in GERD patients with PPI use
A. Cladogram derived from LEfSe analysis of metagenomic sequences of gastric mucosal samples from HCs and GERD patients. The prefixes “p”, “c”, “o”, “f”, and “g” indicate the phylum, class, order, family, and genus, respectively. B. LEfSe comparison of the microbiota in gastric samples from GERD patients with or without PPI use and the HC group. Enriched taxa in samples from GERD patients and HCs with different classification levels with an LDA score >3.0 are shown. C. Extended error bar plots showing functional properties that differ between the gastric mucosal microbiota of non-PPI-users, short-term PPI-users, and long-term PPI-users. LEfSe, linear discriminant effect size; LDA, linear discriminant analysis.