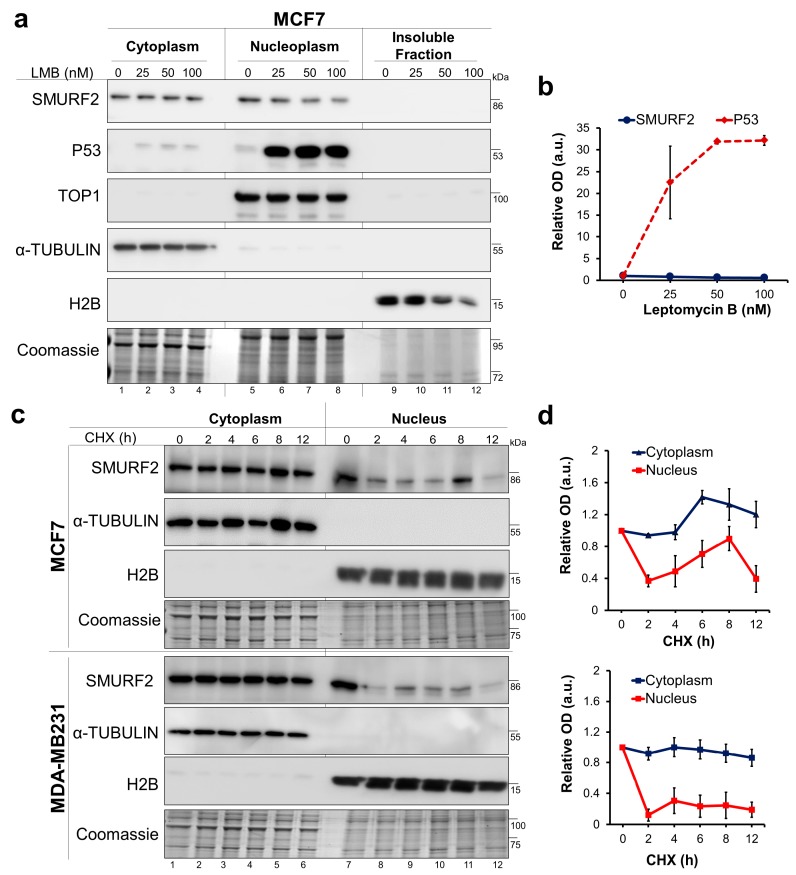
Figure 5.
Mechanistic aspects associated with the distorted biodistribution of SMURF2 in cancer cells. (a) Western blot analysis of SMURF2 and p53 expression and localization in MCF7 cells following cell treatment with CRM1 inhibitor Leptomycin B (LMB; 4 h). α-TUBULIN, TOP1 and H2B were used as controls to validate the equal protein loading and quality of fractionation. Coomassie-gel staining was incorporated into the experiment as an additional control. (b) Quantification of the western blot analysis data of SMURF2 nuclear levels following cell treatment with LMB relative to nuclear loading control TOP1. Data are Mean ± SEM (n = 3). (c) Immunoblot analyses conducted on CHX-treated (50 μg/mL) and subsequently fractionated MCF7 and MDA-MB231 cells show that cytoplasmic and nuclear SMURF2 significantly differ in their protein stabilities. In these experiments, nuclear fractions contained both soluble and insoluble fractions solubilized with sonication. (d) Quantification of the western blot analysis data on SMURF2 turnover levels in the cytoplasmic and nuclear fractions of MCF7 (upper panel) and MDA-MB231 cells (bottom panel) obtained from three independent experiments. SMURF2 cytoplasmic and nuclear levels were quantified relative to the cytoplasmic expressed α-TUBULIN and nuclear histone H2B loading controls. Data are Mean ± SEM (n = 3).