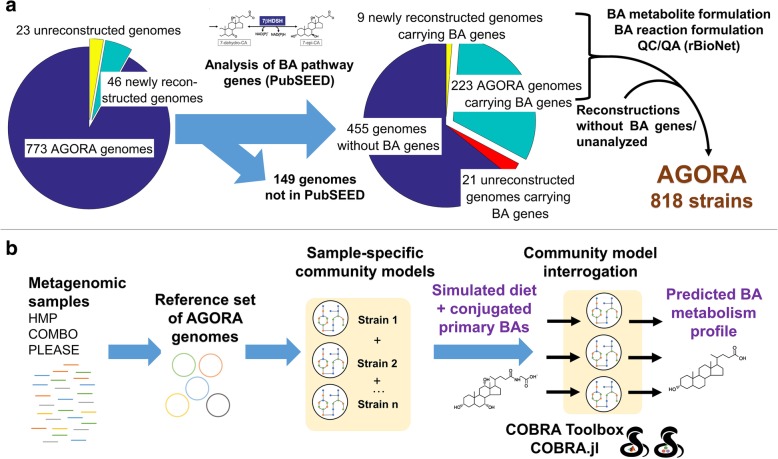
Fig. 1.
Schematic overview of the workflow in this study. a Comparative genomic and metabolic reconstruction approach used to expand the AGORA [15] resource with a bile acid (BA) deconjugation and biotransformation subsystem. The comparative genomic approach was performed in the PubSEED [26, 27] platform. Quality controland quality assurance (QC/QA) during reaction and metabolite formulation and addition to the AGORA reconstructions were ensured by using the reconstruction tool rBioNet [74]. b Computational pipeline used to predict the sample-specific bile acid deconjugation and biotransformation by human gut microbiomes. First, publicly available metagenomic data was retrieved from HMP [35], and the COMBO/PLEASE [36, 37] cohort. Next, the strain-level abundances were mapped onto the reference set of AGORA genomes. Microbial community models were constructed using the illustrated workflow, as implemented in the Microbiome Modeling Toolbox [33], and they account for the strain-level composition of each individual microbiome. Finally, each personalized community model was constrained with an “Average European” diet supplemented with conjugated primary bile acids and its individual-specific, primary bile acid deconjugation and biotransformation potential was computed using flux balance analysis [9, 76].