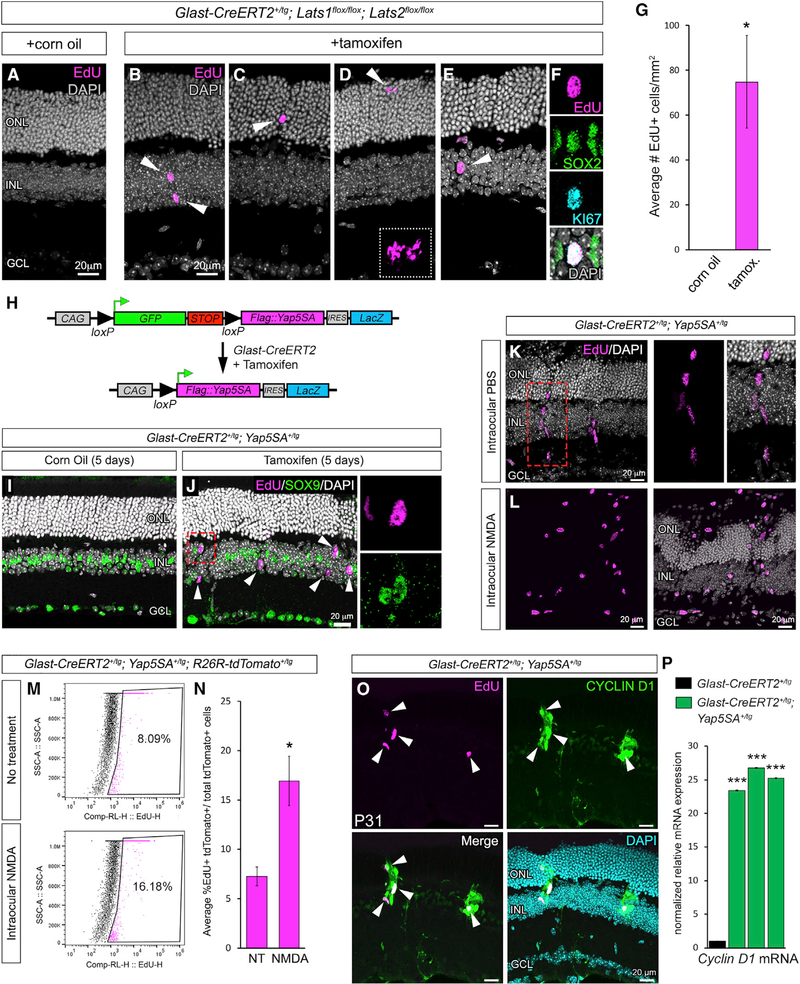
Figure 3. Genetic Loss or Bypass of Hippo Signaling Results in Spontaneous MG Proliferation.
(A–D) EdU labeling (magenta) of Lats1/Lats2 on control (A) and CKO (B, C, and D) retinae.
(E and F) EdU labeling (magenta) and immunofluorescence for SOX2 (green) and KI67 (cyan) in CKO (E and F). (F) shows the same nuclei as in (E) (arrowhead).
(G) Quantification of EdU+ cells in tamoxifen-injected mice versus corn oil vehicle controls.
(H) Map of the Yap5SA transgene.
(I and J) EdU labeling (magenta) and SOX9 (green) immunofluorescence on control (I) and YAP5SA-expressing (J) retinae (the boxed nucleus in J is shown in the insets).
(K) EdU labeling of radially oriented nuclear clusters.
(L) EdU labeling of YAP5SA+ retinae that were also exposed to NMDA damage.
(M and N) Flow cytometry (M) and quantification (N) of EdU-labeled, tdTomato+ cells from untreated and NMDA-damaged retinae expressing YAP5SA.
(O) CYCLIN D1 immunofluorescence and EdU labeling of YAP5SA+ retinae.
(P) Cyclin D1 qRT-PCR from three independent pools of tamoxifen-induced Glast-CreERT2+/tg; Yap5SA+/tg retinae (green bars) compared to tamoxifen-induced Glast-CreERT2+/tg retinae (black bar).
For qRT-PCR, mRNA levels are relative to the control (set to 1) and depicted as fold change ± SEM (n = 3 independent pooled samples per group; Student’s t test). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.