Abstract
Studies have rarely been conducted on the role of miRNAs in prostate cancer (PCa) cell progression by directly targeting MTDH, at least to the best of our knowledge. Thus, the present study aimed to identify miRNAs closely related with metadherin (MTDH) and to determine their roles in PCa. For this purpose, the expression levels of MTDH in PCa tissues and cell lines were examined by RT-qPCR, immunohistochemistry and western blot analysis. By cell transfection, MTDH was either overexpressed in the normal prostate epithelial cell lines or silenced in tumor cell lines to determine cell viability, invasion and migration. Bioinformatics analysis, RT-qPCR, western blot analysis, dual-luciferase reporter assay and MTT assay were performed to identify direct the target of MTDH and to examine tumor cell viability. Rescue experiments using the PC-3 and LNCaP cells were carried out by MTT assay, scratch wound assay, Transwell assay, RT-qPCR and western blot analysis. Experiments were also conducted using 46 PCa human cancer and adjacent tissues, as wells as on 501 cases of PCa from the TCGA database. It was confirmed that the overexpression of MTDH was associated with a poor prognosis of patients. The overexpression of MTDH was found to promote the viability, invasion and migration of PCa cells. miR-145-5p and miR-145-3p identified from 16 miRNAs were found to be closely related to PCa and to be the targets of MTDH. Both these miRNAs were found to significantly suppress the growth and metastasis of PCa cells by negatively regulating the expression of MTDH. On the whole, the findings of this study demonstrate that MTDH functions as an oncogene in PCa and the inhibition of MTDH by miR-145-5p or miR-145-3p suppressed the growth and metastasis of PCa cells. The miR-145-5p/MTDH and miR-145-3p/MTDH pathways may thus become novel treatment targets for PCa.
Keywords: metadherin, miR-145-5p, miR-145-3p, prostate cancer, growth, metastasis
Introduction
In the United States, prostate cancer (PCa) is one of the most common malignant tumors affecting males. Almost 161,360 new cases were diagnosed in 2017, leading to 26,730 deaths within one year (1). Although early diagnosis and treatment have reduced the mortality rate of patients with PCa, it is still the second leading cause of cancer-related mortality in the United States among males (1). Radiotherapy and radical prostatectomy are used in organ-confined PCa; however, >40% of patients develop recurrence or metastasis (2). In fact, the majority of PCa cases are diagnosed in the advanced stages of the disease. Androgen deprivation therapy can treat PCa with controlled or improved conditions in a large number of patients; however, after a median remission period of 18-36 months, tumors also recur, gaining the androgen-independent ability to proliferate (3,4). The occurrence and development of tumors is the result of multi-gene and multi-factor actions. Thus, the exploration of the molecular mechanisms and biomarkers of tumorigenesis has attracted much research attention in oncology.
The Human Genome Project has revealed that there is a large amount of non-coding RNAs (ncRNAs) in the human genome, which has been proven by >40-60% of genome transcripts (5). MicroRNAs (miRNAs or miRs) are endogenous small ncRNA molecules (19-22 nucleotides in length) that regulate protein coding. They regulate gene expression by binding to a specific sequence of messenger RNA, thereby inhibiting the translation or shearing of RNA transcripts (6,7). Previous bioinformatics analyses have revealed that miRNAs regulate the expression levels of 60% of all genes (8,9). Therefore, miRNAs are considered to be a factor widely involved in the micro-regulation of gene expression and may participate in almost all biological processes (10,11). In 2002, it was reported that the expression of miRNAs was downregulated in the majority of chronic B-lymphoblastic leukemia tissues, emphasizing for the first time the importance of miRNAs in human cancer (12). Increasing evidence has indicated that abnormally expressed miRNAs regulate tumorigenesis, progression, metastasis and drug resistance by targeting some tumor-related genes (13,14). In prostate cells, some miRNAs are differentially expressed, of which miR-375, miR-200c, miR-143 and miR-145 have been reported to have a particularly significant effect (15-19).
Metadherin (MTDH) is alternatively known as astrocyte elevated gene (AEG)-1 (20,21). A number of studies have found that MTDH is highly expressed in several tumor cells and is closely related to the proliferation, apoptosis and migration of tumor cells (22-26). There is evidence to indicate that several miRNAs are involved in tumor cell progression by directly targeting MTDH (27-30).
Although, several studies had investigated the effects of MTDH or other miRNAs targeting MTDH on PCa, an miRNA can regulate multiple mRNA targets, and similarly, multiple miRNAs can act on the same mRNA (31,32). Moreover, as a heterogeneous tumor, on the one hand, PCa exhibits an abnormal miRNA expression; for instance, miR-183 has been found to be upregulated in PCa, while miR-miR-145 has been shown to be downregulated in prostate tissues (17). On the other hand, MTDH may have different functions in different PCa cell lines. It can be thus concluded that an individual miRNA may play different roles in different cell types by targeting different pathways/genes. Therefore, it is still necessary to study the relations among new miRNAs, MTDH and PCa, as well as their effects on tumor growth and metastasis.
Materials and methods
Tissue samples
A total of 46 PCa tissues and adjacent normal tissues were collected from patients with PCa who underwent resection surgery and were admitted to the First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology (Luoyang, China) from June, 2011 to May, 2012. Tissues were confirmed by pathological analysis. One part of the paired tissues were used for pathological diagnosis and stored in 4% formaldehyde solution, while the other part was used in later experiments and stored in a liquid nitrogen tank. The association between MTDH expression and the clinicopathological characteristics of patients with PCa is presented in Table I. The Ethics Committees of the First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology approved the study. All patients signed informed consent forms prior to participation.
Table I.
The association between MTDH expression and the clinicopathological characteristics of patients with prostate cancer.
| Characteristics | No. of patients (n=46) | No. of patients with high MTDH expression (n=31) | No. of patients with low MTDH expression (n=15) | P-value |
|---|---|---|---|---|
| Age (years) | 0.592b | |||
| <55 | 21 | 15 | 6 | |
| ≥55 | 25 | 16 | 9 | |
| Median survival | 26.3 | 25.4 | 28.4 | 0.468 |
| Tumor size (cm) | 0.047a,b | |||
| <3 | 21 | 11 | 10 | |
| ≥3 | 25 | 20 | 5 | |
| TNM stage | 0.497c | |||
| I and II | 33 | 21 | 12 | |
| IIIa | 13 | 10 | 3 | |
| Osseous metastasis | 0.027a,b | |||
| No | 20 | 10 | 10 | |
| Yes | 26 | 21 | 5 | |
| Lymphatic metastasis | 0.161c | |||
| No | 27 | 16 | 11 | |
| Yes | 19 | 15 | 4 |
P<0.05, statistical significance as shown by the Chi-square test (b) or Fisher’s test (c).
Immunohistochemistry (IHC)
IHC was performed to detect the protein expression levels of MTDH in 2 paired PCa and adjacent tissues by streptavidin-peroxidase (SP) staining. The fresh PCa and adjacent tissues were fixed in 4% formaldehyde for >24 h at room temperature. The samples (5 μm thickness) were deparaffinized in xylene, dehydrated with gradient ethanol, and 3% H2O2 was used to block endogenous peroxidase. The samples were first incubated with MTDH antibody (1:2,000; ab227981; Abcam, Cambridge, MA, USA) at 4°C overnight and then were washed by PBS and incubated at room temperature for 30 min with the secondary antibody, HRP-conjugated goat anti-rabbit IgG (1:1,000; cat. no. 10285-1-AP; Proteintech Group, Inc., Rosemont, IL, USA). Diaminobenzidine (DBA) was performed by chromogen and hematoxylin was used to re-dye the samples. The substitution of PBS for primary antibody was used as a negative control. Staining patterns were analyzed by selected representative slices. The immunostains were observed by 2 independent experienced pathologists. MTDH staining intensities were categorized as negative, faint yellow, yellow and brown.
Cells and cell culture
Human prostate epithelial cell lines [PWR-1E (CRL-11611) and RWPE-1 cells (CRL-11609)] and PCa cell lines [PC-3 (CRL-1435™), DU145 (HTB-81), C4-2 (CRL-3314), 22Rv1 (CRL-2505) and NCI-H660 cells (CRL-5813)] were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and LNCaP cells were purchased from Cell Lines Service (CLS, Eppelheim, Germany). The cells were cultured at 37°C in 5% CO2 in RPMI-1640 medium (GENOM, Hangzhou, China) with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco/Thermo Fisher Scientific). The medium was changed every 8-10 h.
Cell transfection
Cells were seeded in a 6-well plate (1.0×105) for 24 h prior to transfection. Overexpression and MTDH siRNA (silencing MTDH, 5′-GGTGAAGATAACTCTACTG-3′), inhibitors and mimics of miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p, as well as mock and negative control (NC, 5′-GGACGAUGGCUAAUUACAU-3′) plasmids were synthetized by Invitrogen/Thermo Fisher Scientific. Transient transfection was performed using Lipofectamine 2000 (Invitrogen/Thermo Fisher Scientific) according to the manufacturer’s instructions. A total of (100 nmol/l) overexpression or silencing MTDH/ mimics or inhibitors of miRNAs, mock or NC and Lipofectamine 2000 were added to Opti-MEM medium followed by incubation at 25°C for 10 min, respectively. Lipofectamine 2000 was then mixed into each well and cultured in Opti-MEM RPMI-1640 medium. Following 6 h of culturing, the fluid was changed back to RPMI-1640 medium containing 10% FBS.
Cell viability
3-(4,5-Dimethylthiazol-2-yl)-2,5-dipheny-ltetrazolium bromide (MTT; Solarbio Life Sciences, Beijing, China) was used to detect cell viability. The cells (1×104 cells/well) were seeded in 96-well plates and cultured for 24, 48 72 h at 37°C. The cells were then washed with PBS buffer and 20 μl MTT reagent were added to each well and cultured for a further 4 h. Subsequently, 150 μl DMSO were added after the MTT was removed. The optical density was measured at 562 nm using a microplate reader (Thermo Fisher Scientific).
Cell scratch wound assay
The cells were seeded in 6-well plates and incubated at 37°C for 24 h. A wound was drawn in the center of the plate using the sterile 100 μl pipette tip, and PBS was used to gently wash the cells 3 times and serum-free medium was then added. Cell migrations were observed using an inverted microscope (CKX53, Olympus, Tokyo, Japan) per 24 h. The scratch area was measured using ImageJ software Version 1.49 (NIH, Bethesda, MD, USA).
Cell invasion assay
Cell invasion was performed by Matrigel-coated Transwell cell culture chambers. Following transfection, the cells were resuspended in serum-free medium and 1×104 cells were added to the upper chamber coated with Matrigel. In the lower 24-well chamber, DMEM medium containing 10% fetal bovine serum was added and the cells were first incubated for 24 h for 37°C and were then fixed with 1% formaldehyde for 10 min at 25°C and finally stained with 0.5% crystal violet (Leagene Biotechnology, Beijing, China) for a further 5 min at room temperature. Invasion cells were counted at ×200 magnification.
Bioinformatics analysis
Data regarding 501 cases of PCa were downloaded from the TCGA database. The association between MTDH and overall survival was detected by the Kaplan-Meier method, followed bythe log rank test. Differential expression levels of miRNAs in normal prostate tissues and cancer tissues were screened by DianaTools (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=site/page&view=software). DianaTools, miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php), miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) and TargetScan (http://www.targetscan.org/vert_72/) were used to predict the miRNAs targeting MTDH.
Luciferase assay
Empty vector or miR-145-5p/miR-145-3p/ miR-499a-5p/miR-22-3p and luciferase reporter comprising 3′-UTR of MTDH wild-type or mutant fragment (GeneChem, Shanghai, China) were co-transfected using Lipofectamine 2000 (Invitrogen/Thermo Fisher Scientific) into 293T cells (ACS-4500; ATCC) cultured in a 96-well plate. Following 48 h of transfection, the cells were harvested and luciferase activity was measured the chemiluminescence using the dual-luciferase reporter assay (normalization with Renilla luciferase activity, Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Reverse transcription-quantitative PCR (RT-qPCR)
According to the manufacturer’s instructions, total RNA was isolated from the tissues or cells using TRIzol regent (Invitrogen/Thermo Fisher Scientific). The GoScriptTM Reverse Transcription kit (Promega) was used for reverse transcription at 37°C for 60 min and at 85°C for 5 min. qPCR was carried out on SYBR Fast qPCR Mix (Invitrogen/Thermo Fisher Scientific) for MTDH, hsa-miR-145-5p, hsa-miR-145-3p, hsa-m iR- 499a-5p, hsa-m iR-22-3p, hsa-m iR-375, hsa-miR-30e-5p, hsa-miR-30a-5p, hsa-miR-200b-3p, hsa-miR-34b-5p, hsa-miR-217, hsa-miR-378a-5p, hsa-miR-200c-3p, hsa-miR-136-5p, hsa-miR-30c-5p, hsa-miR-320a and hsa-miR-30b-5p. The primer sequences are listed in Table II. Samples were run using the following cycling parameters: 95°C for 5 min, 95°C for 10 sec, followed by 40 cycles of 60°C for 20 sec and 72°C for 10 sec. After completion of the PCR amplification, the 2-ΔΔCq [relative quantity (RQ)] method was used to detect comparative quantification (33). Primers were synthetized commercially (Invitrogen/Thermo Fisher Scientific).
Table II.
Primers used in RT-qPCR.
| Gene | Primer | Sequence |
|---|---|---|
| MTDH | Forward | 5′-TGCCTCCTTCACAGACCAA-3′ |
| Reverse | 5′-TCGGCTGCAGATGAGATAG-3′ | |
| hsa-miR-145-5p | Forward | 5′-GTCCAGTTTTCCCAGGAATCCCT-3′ |
| Reverse | 5′-TGGTGTCGTGGAGTCG-3′ | |
| hsa-miR-145-3p | Forward | 5′-GCCCTGTAGTGTTTCCTACTT-3′ |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-499a-5p | Forward | 5′-GACTTGCAGTGATGTTTGTCGT-3′ |
| Reverse | 5′-TGTCGTGGAGTCGGCAATTG-3′ | |
| hsa-miR-22-3p | Forward | 5′-AAGCTGCCAGTTGAAGAACTGTA-3′ |
| Reverse | 5′-GCTGTCAACGATACGCTACGTAAC-3′ | |
| hsa-miR-375 | Forward | 5′-TTGTTCGTTCGGCTCGCG-3′ |
| Reverse | 5′-TTTGGCACTAGCACATT-3′ | |
| hsa-miR-30e-5p | Forward | 5′-GGCGTGTAAACATCCTTGACTG-3′ |
| Reverse | 5′-GTGCAGGGTCCGAGGT-3′ | |
| hsa-miR-30a-5p | Forward | 5′-ACATCCTCGACTGGAAGGTC-3′ |
| Reverse | 5′-TGTCGTGGAGTCGGCAATTG-3′ | |
| hsa-miR-200b-3p | Forward | 5′-TGCCTGGTAATGATGAGTCGT-3′ |
| Reverse | 5′-GTGTCGTGGAGTCGGCAATT-3′ | |
| hsa-miR-34b-5p | Forward | 5′-TGATTGGTCGTATCCAGTGCAA-3′ |
| Reverse | 5′-GTATCCAGTGCGTGTCGTGG-3′ | |
| hsa-miR-217 | Forward | 5′-TCCTAATGCATTGCCTTCAGC-3′ |
| Reverse | 5′-CGGCAATTGCACTGGATACG-3′ | |
| hsa-miR-378a-5p | Forward | 5′-TCCAGGTCCTGTGTGTCGTA-3′ |
| Reverse | 5′-GTATCCAGTGCGTGTCGTGG-3′ | |
| hsa-miR-200c-3p | Forward | 5′-TCGTCTTACCCAGCAGTG-3′ |
| Reverse | 5′-CGGCAGTATTAGAGACTCC-3′ | |
| hsa-miR-136-5p | Forward | 5′-TGGAGTCGTATCCAGTGCAA-3′ |
| Reverse | 5′-GTCGTATCCAGTGCGTGTCG-3′ | |
| hsa-miR-30c-5p | Forward | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| Reverse | 5′-CGCTTCACGAATTTGCGTGTCAT-3′ | |
| hsa-miR-320a | Forward | 5′-ATCCAGTGCAGGGTCCGAGG-3′ |
| Reverse | 5′-CGCGGTTAAAAGCTGGGTTGAGA-3′ | |
| hsa-miR-30b-5p | Forward | 5′-AGCTGTCGTATCCAGTGCAA-3′ |
| Reverse | 5′-GTCGTATCCAGTGCGTGTCG-3′ | |
| β-actin | Forward | 5′-CATGTACGTTGCTATCCAGGC-3′ |
| Reverse | 5′-CTCCTTAATGTCACGCACGA-3′ |
Western blot analysis
RIPA lysis buffer (Beyotime, Shanghai, China) was used to extract the protein, which was obtained from the tissues and cells. The concentration of the proteins was detected using a BCA protein kit (Beyotime). Aliquots of protein were separated by 12% SDS-PAGE and resolved proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA), which were blocked in 5% milk PBS with 0.1% Triton X-100 and incubated with anti-MTDH primary antibody (1:1,000; ab227981; Abcam) overnight at 4℃. The membranes were then incubated with the appropriate HRP-conjugated secondary antibody (1:10,000; cat. no. 10285-1-AP; Proteintech). Protein bands were detected with ECL (Thermo Fisher Scientific) and visualized using Quantity One software Version 4.6.2 (Bio-Rad, Hercules, CA, USA).
Statistical analysis
GraphPad Prism software version 6.0 was used to conduct statistical analysis. All data are presented as the means ± standard deviation (SD). Differences were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. The Chi-square test or Fisher’s test were carried out to examine the association between MTDH expression and the clinicopathological characteristics of the patients with PCa. The cut-off value of the patients with a high MTDH expression was >1(RQ) and the cut-off value of patients with a low MTDH expression was <1(RQ). A value of P<0.05 was considered to indicate a statistically significant difference.
Results
MTDH is upregulated in PCa tissues and is associated with a poor prognosis of patients with the disease
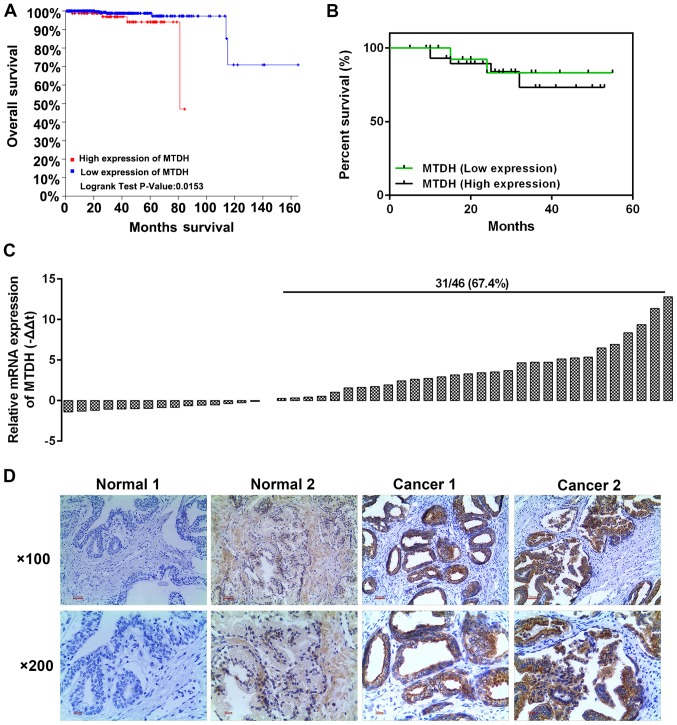
The analysis of 501 PCa cases from the TCGA database revealed that a high expression of MTDH was associated with a relatively low overall survival rate (P=0.0153; Fig. 1A). We selected 46 cases of PCa tissues and adjacent normal tissues. The results of RT-qPCR revealed that a high expression of MTDH existed 31 PCa tissues (Fig. 1C). Kaplan-Meier analysis revealed that the patients with a high expression of MTDH had a poor 5-year survival rate (Fig. 1B). Furthermore, immunohistochemical staining was performed to determine protein expression in two randomly selected patients. As shown in Fig. 1D, the selected patient tissues were representative in a way that, MTDH had high expression in the tumor tissues with a few brown granules located in the nucleus and the majority of brown granules were confined to the cytoplasm and cell membrane (Fig. 1D). However, the normal tissues had a negative or weak expression (resembling the color of popcorn) of MTDH. Furthermore, the analysis of the association between MTDH expression and the clinicopathological characteristics of the patients with PCa revealed that MTDH expression was significantly associated with tumor size and osseous metastasis (P<0.05; Table I). The median survival in both groups exhibited no significant difference, and such a phenomenon may be explained by the fact that the sample size was not large enough.
Figure 1.
Expression of MTDH in prostate cancer (PCa) tissues and adjacent normal tissues, and its association with patient prognosis. (A) The TCGA database was used to analyze the association between MTDH and overall survival. (B) A total of 46 PCa and adjacent normal tissues were detected to determine the association between MTDH and overall survival by Kaplan-Meier analysis. (C) A total of 46 PCa tissues were detected for their mRNA expression of MTDH by RT-qPCR. (D) Immunohistochemistry was used to obverse the protein level of MTDH in two randomly selected patients (magnification: Upper panels, ×100; lower panels, ×200). MTDH, metadherin.
Expression of MTDH in normal prostate and PCa cell lines
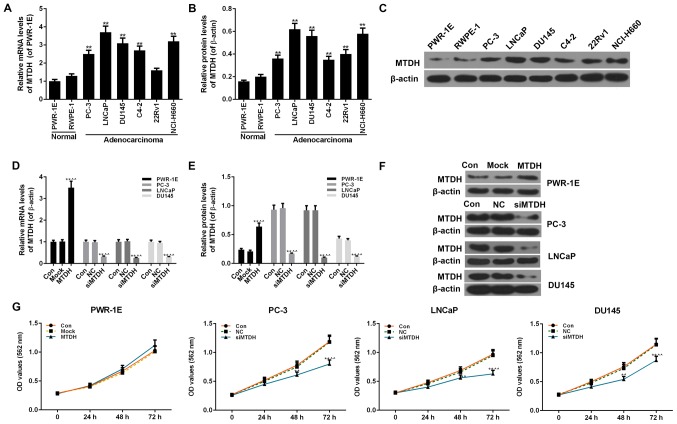
Two normal human prostate epithelial cell lines (PWR-1E and RWPE-1 cells) and 6 PCa cell lines (PC-3, LNCaP, DU145, C4-2, 22Rv1 and NCI-H660 cells) were examined to determine the expression levels of MTDH. The 2 normal cells exhibited no obvious differences in the mRNA and protein expression of MTDH. However, compared with the 2 normal cells, almost all 6 PCa cell lines exhibited a significantly increased expression of MTDH at both the mRNA and protein level (P<0.01; Fig. 2A-C).
Figure 2.
Expression of MTDH in prostate cancer (PCa) and normal cell lines, and the effect of the overexpression or silencing MTDH on cell viability. (A) RT-qPCR was used to detect the mRNA level of MTDH in 6 PCa (PC-3, LNCaP, DU145, C4-2, 22Rv1 and Ncl-H660 cells) and 2 normal cell lines (PWR-1E and RWPE-1 cells). (B and C) Western blot analysis was used to detect the protein level of MTDH in 6 PCa (PC-3, LNCaP, DU145, C4-2, 22Rv1 and Ncl-H660 cells) and 2 normal cell lines (PWR-1E and RWPE-1 cells). Relative protein levels of MTDH are shown as bar diagrams (**P<0.01, compared with normal cell lines). The expression of each protein in cells was determined following normalization with a loading control, β-actin. (D-F) Transfection efficiency of MTDH in prostate normal and cancer cell lines following transfection with MTDH overexpression plasmid or MTDH siRNA, both at the (D) mRNA and (E and F) protein levels determined by RT-qPCR and western blot analysis. β-actin served as an internal control. (G) MTT assay was performed to determine the effects of the overexpression/silencing MTDH on cell viability at 24, 48 and 72 h [*P<0.05 and **P<0.01, compared with control; ^P<0.05 and ^^P<0.01, compared with mock/negative control (NC) groups]. Data are shown as the means ± SD from 3 independent experiments. MTDH, metadherin.
Effects of the overexpression or silencing of MTDH on cell viability, and on the invasion and migration of normal and PCa cell lines
The overexpression of MTDH was successfully induced in the normal human prostate epithelial cell line, PWR-1E, by transfection with an MTDH overexpression plasmid. The results of RT-qPCR and western blot analysis revealed that the expression of MTDH significantly increased in the PWR-1E cells by transfection with MTDH overexpression plasmid, compared to the control (P<0.01; Fig. 2D-F). In addition, 3 PCa cell lines (PC-3, LNCaP and DU145) that had a higher expression of MTDH or studied widely were transfected with siRNA against MTDH (siMTDH). No differences were observed between the control and NC groups in terms of the expression of MTDH. However, in the siMTDH groups, the mRNA and protein levels of MTDH exhibited a significant decrease (P<0.01; Fig. 2D-F). As regards cell viability, the overexpression of MTDH slightly enhanced the viability of the normal PWR-1E cells in comparison to the control or mock groups (P>0.05; Fig. 2G). In the PC-3, LNCaP and DU145 PCa cells, the silencing of MTDH significantly decreased cell viability starting from 48 h in the siMTDH groups (48 h, P<0.05; 72 h, P<0.01; Fig. 2G).
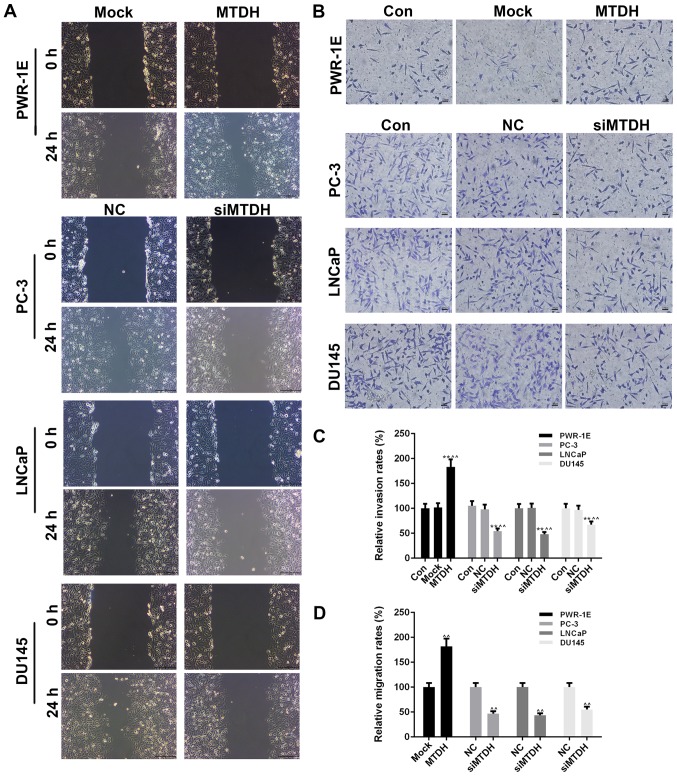
Cell migration and invasion were also detected by scratch wound assay and Transwell assay, respectively (Fig. 3A and B). At 24 h after the scratch wound was created, the overexpression of MTDH significantly increased the migration of the PWR-1E cells (P<0.01; Fig. 3D), while the silencing of MTDH significantly decreased the migration of the PCa cells (P<0.01). The effects of the overexpression or silencing of MTDH on cell invasion were similar to those observed on cell migration. Specifically, the overexpression of MTDH increased the invasion of the PWR-1E, while the silencing of MTDH decreased the invasion of the 3 PCa cell lines (P<0.01; Fig. 3C).
Figure 3.
Effects of overexpressing or silencing MTDH on cell invasion and migration in prostate normal and cancer cell lines. (A) Cell scratch wound assay was carried out to detect the effects of the overexpression or silencing of MTDH on the migration of the normal PWR-1E cells or 3 prostate cancer (PCa) cell lines (PC-3, LNCaP and DU145 cells). (B) Transwell assay was performed to detect the effects of the overexpression or silencing of MTDH on invasion of PWR-1E normal cells or 3 PCa cell lines (PC-3, LNCaP and DU145 cells). (C) Relative invasion rates are shown as bar diagrams. (D) Relative migration rates are shown as bar diagrams. Data are shown as the means ± SD from 3 independent experiments [**P<0.01, compared with control; ^^P<0.01, compared with mock/negative control (NC) groups]. MTDH, metadherin.
Screening of miRNAs associated with MTDH and the expression of miRNAs in normal prostate normal and PCa cell lines
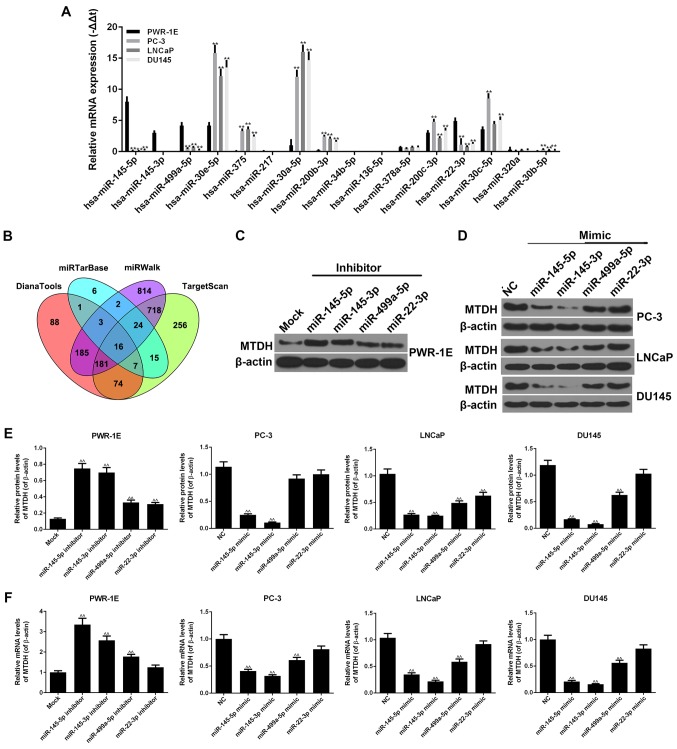
By using the DianaTools, TargetScan, miRWalk and miRTarBase databases, 16 miRNAs were found to be associated with MTDH in PCa and normal tissues (Fig. 4B). As shown by RT-qPCR, similar results were observed for hsa-miR-145-5p, hsa-miR-145-3p, hsa-miR-499a-5p and hsa-miR-22-3p, in that they exhibited high expression levels in the normal PWR-1E cells, and relatively low expression levels in the PC-3, LNCaP and DU145 PCa cells (P<0.01; Fig. 4A). Similar results were also observed for hsa-miR-30e-5p, hsa-miR-375, hsa-miR-30a-5p, hsa-miR-200b-3p, hsa-miR-200c-3p, hsa-miR-30c-5p and hsa-miR-30b-5p, in that they exhibited high expression levels in the PC-3, LNCaP and DU145 cells, but low expression levels in the PWR-1E cells (P<0.01). The expression levels of other miRNAs did not exhibit clear differences between the normal and cancer cell lines.
Figure 4.
miRNAs are closely associated with MTDH. (A) RT-qPCR was used to detect the expression levels of 16 miRNAs in PWR-1E, PC-3, LNCaP and DU145 cells. (B) The 16 miRNAs associated with MTDH were screened by DianaTools, miRTarBase, miRWalk and TargetScan (**P<0.01, compared to normal PWR-1E cells). (C) Four representative miRNAs, miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p, were selected to perform the following experiments. The inhibitors of these 4 miRNAs were used to determine their effects on the protein expression of MTDH in PWR-1E cells. β-actin served as an internal control. (D) The mimics of these 4 miRNAs were used to determine their effects on the protein expression of MTDH in PC-3, LNCaP and DU145 cells. β-actin served as an internal control. (E) The effects of the inhibitors or mimics of the 4 miRNAs at the protein levels of MTDH in PWR-1E or 3 prostate cancer (PCa) cells are shown as bar diagrams. (F) The effects of the inhibitors or mimics of the 4 miRNAs on the mRNA levels of MTDH in PWR-1E or 3 PCa cells are shown as bar diagrams. Data are shown as the means ± SD from 3 independent experiments [^^P<0.01, compared to mock/negative control (NC) group]. MTDH, metadherin.
Effects of inhibitors or mimics of miRNAs on the expression of MTDH
We selected 4 representative miRNAs, miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p, which had a high expression in the PWR-1E cells, but a low expression in the PC-3, LNCaP and DU145 cells. The inhibitors of these 4 miRNAs were transfected into the PWR-1E cells. The results revealed that all 4 miRNA inhibitors increased the protein and mRNA expression levels of MTDH (Fig. 4C, E and F). In particular, the increasing effects of the miR-145-5p and miR-145-3p inhibitors on the expression of MTDH were more significant than those of the other inhibitors (P<0.01). miR-22-3p inhibitor exerted a comparative effect on the mRNA expression of MTDH in comparison with the mock group. In addition, the mimics of these 4 miRNAs were transfected into 3 PCa cell lines. The miR-145-5p and miR-145-3p mimics noticeably decreased the expression of MTDH at both the protein and mRNA level (P<0.01; Fig. 4D-F). The miR-22-3p mimic hardly inhibited the expression of MTDH in the PCa cells, apart from the LNCaP cells. In the PC-3, LNCaP and DU145 cell lines, the miR-499a-5p mimic decreased the expression of MTDH (P<0.01), although not at the protein level in the PC-3 cells.
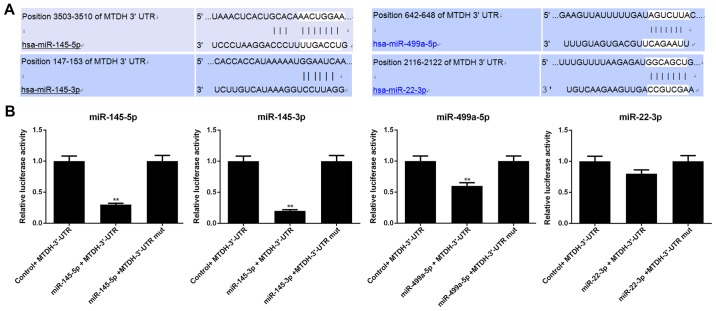
MTDH is a direct target of miR-145-5p, miR-145-3p and miR-499a-5p
Luciferase reporter assay was conducted to determine whether MTDH is regulated by miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p. Luciferase reporter was subcloned with the wild-type or mutant sequence of MTDH and was then co-transfected with miR-145-5p, miR-145-3p, miR-499a-5p, miR-22-3p or miR-NC into 293T cells (Fig. 5). The luciferase activities significantly decreased after miR-145-5p/ miR-145-3p/miR-499a-5p and MTDH 3′-UTR-wt were co-transfected into the 293T cells (P<0.01); however, the luciferase activities in the cells co-transfected with MTDH 3′-UTR-mut and miR-145-5p/miR-145-3p/miR-499a-5p (P>0.05) remained stable. In addition, neither MTDH 3′-UTR-wt nor MTDH 3′-UTR-mut with miR-22-3p exerted obvious effects on luciferase activity (P>0.05).
Figure 5.
Verification of whether MTDH is a direct target of miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p. (A) Putative miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p binding sites in the 3′-UTR of MTDH. (B) Relative luciferase activity in 293T cells following co-transfection with wild-type or mutant (mut) MTDH 3′-UTR reporter, which was constructed together with miR-145-5p, miR-145-3p, miR-499a-5p or miR-22-3p (**P<0.01, compared with control + MTDH-3′-UTR group).
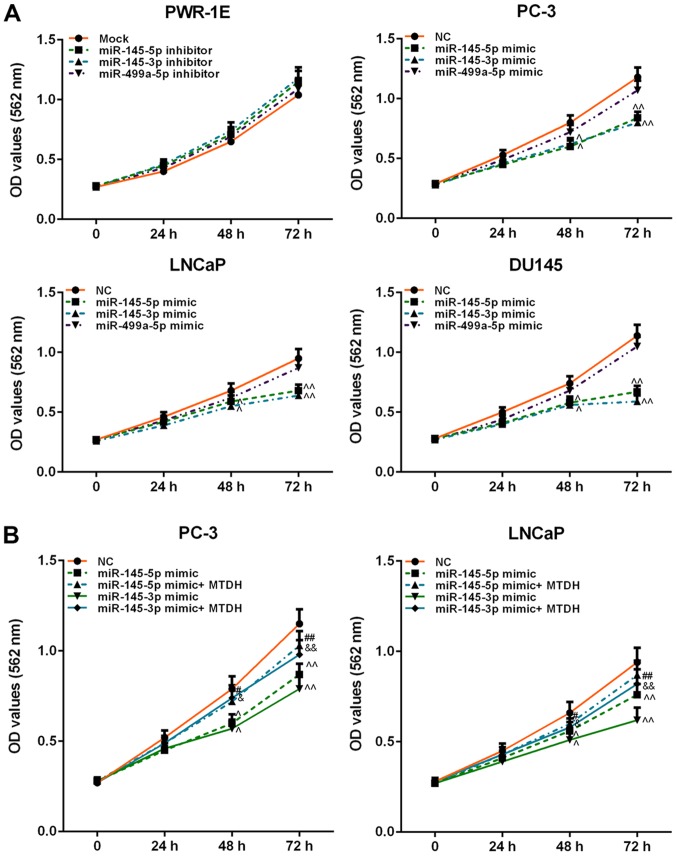
Effects of inhibitors or mimics of miR-145-5p/miR-145-3p/ miR-499a-5p on cell viability
In order to determine the effects of miR-145-5p/miR-145-3p/miR-499a-5p on normal prostate or PCa cell viability, inhibitors or mimics were used to determine the effects on cell viability. The result revealed that inhibitors of these 3 miRNAs weakly increased PWR-1E cell viability (P>0.05; Fig. 6A). However, the mimics of miR-145-5p and miR-145-3p exerted a similar effect on PCa cell viability, which was significantly decreased at 48 h (P<0.05). miR-499a-5p slightly attenuated the viability of the PC-3, LNCaP and DU145 cells (P>0.05; Fig. 6A).
Figure 6.
Effects of inhibitors or mimics of miR-145-5p, miR-145-3p and miR-499a-5p, as well as in combination with MTDH on cell viability. (A) Effects of inhibitors or mimics of miR-145-5p, miR-145-3p and miR-499a-5p on PWR-1E or PC-3, LNCaP, DU145 cell viability, detected by MTT assay. (B) Effects of MTDH on decreasing cell viability induced by miR-145-5p/ miR-145-3p mimic in PC-3 and LNCaP cells. Data are shown as the means ± SD from 3 independent experiments [^P<0.05 and ^^P<0.01, compared with mock/negative control (NC); #P<0.05 and ##P<0.01, compared with miR-145-5p mimic; &P<0.05 and &&P<0.01, compared with miR-145-3p mimic]. MTDH, metadherin.
MTDH overexpression reverses the suppressive effects of miR-145-5p and miR-145-3p mimics on the viability, migration and invasion of PCa cells
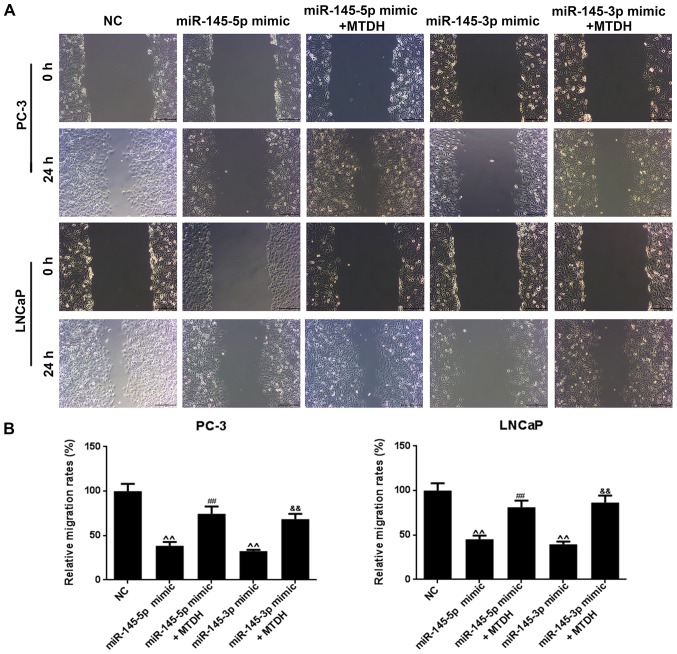
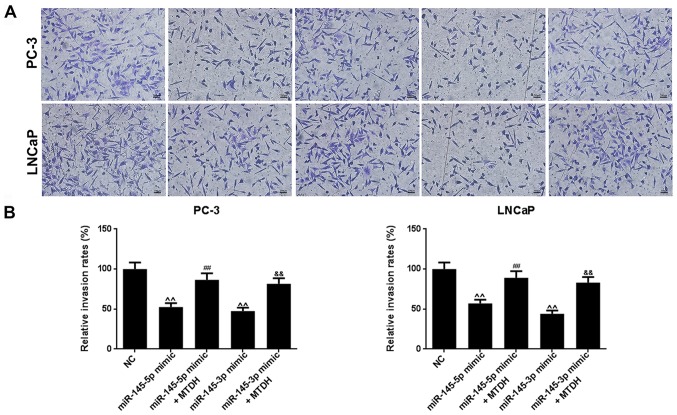
Furthermore, we examined the effects of MTDH in combination with mimics of miR-145-5p/miR-145-3p on PCa cell viability, cell migration and invasion. As shown in Fig. 6B, both miR-145-5p and miR-145-3p mimics significantly decreased PC-3 and LNCaP cell viability at 48 h, compared to the NC group (P<0.05; Fig. 6B). However, when the MTDH overexpression plasmid and miR-145-5p /miR-145-3p mimics were co-transfected into the PC-3 or LNCaP cells, cell viability significantly increased at 72 h in comparison with the group transfected with the mimics alone (P<0.01). As regards cell migration (Fig. 7A), the results revealed that miR-145-5p and miR-145-3p mimics significantly inhibited PC-3 and LNCaP cell migration (P<0.01; Fig. 7B). Similarly, as shown in Fig. 8, miR-145-5p and miR-145-3p mimics significantly inhibited PC-3 and LNCaP cell invasion (P<0.01, Fig. 8). However, the overexpression of MTDH, reversed the inhibitory effects of the 2 miRNAs on migration and invasion (P<0.01).
Figure 7.
MTDH reverses the suppressive effects of miR-145-5p and miR-145-3p mimics on PCa cell migration. (A) Cell migration was examined by cell scratch wound assay in PC-3 and LNCaP cells. (B) The effects of MTDH overexpression on the decreased migration of PC-3 and LNCaP cells induced by miR-145-5p/ miR-145-3p mimics are shown as bar diagrams. Data are shown as the means ± SD from 3 independent experiments [^^P<0.01, compared with mock/negative control (NC); ##P<0.01, compared with miR-145-5p mimic; &&P<0.01, compared with miR-145-3p mimic]. MTDH, metadherin.
Figure 8.
MTDH reverses the suppressive effects of miR-145-5p and miR-145-3p mimics on PCa cell invasion. (A) Cell invasion was examined by Transwell assay in PC-3 and LNCaP cells. (B) The effects of MTDH overexpression on the decreased invasion of PC-3 and LNCaP cells induced by miR-145-5p/miR-145-3p mimics are shown as bar diagrams. Data are shown as the means ± SD from 3 independent experiments [^^P<0.01, compared with mock/negative control (NC); ##P<0.01, compared with miR-145-5p mimic; &&P<0.01, compared with miR-145-3p mimic]. MTDH, metadherin.
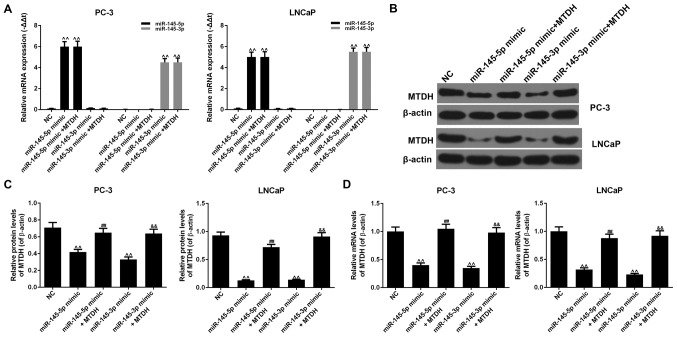
miR-145-5p or miR-145-3p directly target MTDH to negatively regulate its expression
The results revealed that the mRNA expression levels of miR-145-5p or miR-145-3p increased significantly when miR-145-5p or miR-145-3p mimic were respectively transfected into the PC-3 or LNCaP cells (P<0.01; Fig. 9A). The overexpression of MTDH did not affect the expression of miR-145-5p or miR-145-3p compared to the NC group (P<0.01). The results also revealed that miR-145-5p mimic noticeably decreased the protein and mRNA expression levels of MTDH, compared to the NC group in both the PC-3 and LNCaP cells (P<0.01; Fig. 9B-D). However, when miR-145-5p mimic and MTDH were used in combination, the expression of MTDH increased at both the protein and mRNA level (P<0.01). Similar results were also observed with miR-145-3p mimic (P<0.01). Therefore, the suppression of PCa cell growth and metastasis induced by the upregulation of miR-145-5p/miR-145-3p involve the downregulation of MTDH expression.
Figure 9.
The underlying mechanisms of the upregulation of miR-145-5p/miR-145-3p inhibiting prostate cancer cell growth and metastasis. (A) The effects of miR-145-5p/miR-145-3p mimics or the overexpression of MTDH on the expression of miR-145-5p or miR-145-3p were determined by RT-qPCR in PC-3 and LNCaP cells [^^P<0.01, compared with negative control (NC)]. (B) The effects of miR-145-5p/miR-145-3p mimics or the overexpression of MTDH on the protein expression of MTDH were determined by western blot analysis. β-actin served as an internal control. (C) Relative protein levels of MTDH are shown as bar diagrams (^^P<0.01, compared with mock/NC; ##P<0.01, compared with miR-145-5p mimic; &&P<0.01, compared with miR-145-3p mimic). (D) Relative mRNA levels of MTDH are shown as bar diagrams (^^P<0.01, compared with mock/NC; ##P<0.01, compared with miR-145-5p mimic; &&P<0.01, compared with miR-145-3p mimic). Data are shown as the means ± SD from 3 independent experiments.
Discussion
MTDH is a highly conserved protein located at 8q22 (30). It is believed that MTDH may be used not only as an independent molecular marker to determine tumor prognosis, but also a molecular target for antitumor therapy (34,35). Li et al found that the mRNA and protein expression levels of MTDH in breast cancer cells were significantly higher than those in normal cells; their statistical data indicated that the expression of MTDH was associated with the clinical features of breast cancer, including staging, the number of positive lymph nodes and tumor type; therefore, MTDH was considered as an independent prognostic factor (36). In this study, we found that a high expression of MTDH was closely associated with a poor prognosis in 501 PCa cases analyzed from the TCGA database. Thus, we were interested in further exploring the effects of MTDH on PCa. For this purpose, 46 cases of PCa tissues and adjacent normal tissues were collected, and RT-qPCR and IHC revealed that MTDH had a significantly increased expression in the cancer tissues. Moreover, Kaplan-Meier analysis also confirmed that the upregulation of MTDH was related to a poor 5-year survival rate of patients with PCa. The analysis of the association between MTDH expression and the clinicopathological characteristics of patients with PCa revealed that MTDH expression was significantly associated with tumor size and osseous metastasis. Cellular experiments revealed that MTDH had a high expression in several PCa cell lines, but a low expression in 2 normal prostate epithelial cells. Thus, a difference between the mRNA expression level and protein expression level of MTDH in 22RV1 cells was identified, and such a phenomenon may be explained by the participation of other factors, such as transcription factors, small RNA, methylation and acetylation. Studies have also suggested that MTDH may be expected to be a novel target in tumor-targeted therapy as it is overexpressed in various types of tumors, including breast cancer, neuroblastoma, esophageal cancer, cervical cancer, non-small cell lung cancer and hepatocellular carcinoma (36-41). The findings of this study demonstrated that on the one hand, the silencing of MTDH inhibited the viability, migration and invasion of PC-3, LNCaP and DU145 PCa cells; on the other hand, the overexpression of MTDH increased PWR-1E normal cell viability, cell migration and invasion. Some studies, for instance, have proposed that MTDH overexpression can enhance the adhesion of tumor cells and promote tumor metastasis (25,42). Hu et al also found that 8q22 genomic repeated gain significantly accelerated the lung metastasis of cancer cells (43).
miRNAs have recently become a main focus in tumor progression. Studies have demonstrated that some miRNAs are associated with PCa. For example, miR-200c-3p/141-3p, miR-375 and miR-93-5p/106b-5p/25-3p have been frequently found to be upregulated in PCa, whereas the expression levels of miR-221-3p and miR-222-3p in PCa tissues have often been proven to be downregulated (15-17,44-46). In a number of tumor cells, miRNAs such as miR-145, miR-497, miR-1297 and miR-655, have been reported to regulate cell proliferation and invasion by targeting MTDH; however, only a few studies to date have investigated the upstream regulatory mechanisms of MTDH in PCa, at least to the best of our knowledge (27,28,47). In this study, by using the DianaTools, TargetScan, miRWalk and miRTarBase databases, 16 miRNAs were screened to determine whether they were associated with MTDH. As miR-145-5p, miR-145-3p, miR-499a-5p and miR-22-3p were overexpressed in PWR-1E cells and downregulated in the PC-3, LNCap and DU145 cells, these 4 miRNAs were selected to help detect their association with MTDH. Western blot analysis, RT-qPCR and dual-luciferase reporter assay confirmed that miR-145-5p, miR-145-3p and miR-499a-5p bound to MTDH and decreased its expression in PCa cells. MTT assay further revealed that only miR-145-5p and miR-145-3p significantly inhibited tumor cell viability. Previous studies have suggested that miR-145-5p and miR-145-3p, as the dual strands of the miR-145 duplex, play roles in a variety of tumors (48-50). In addition, the two miRNAs can inhibit the proliferation of PCa cells (51,52).
In addition, in this study, we found that MTDH reversed the suppressive effects of miR-145-3p or miR-145-5p mimics on the viability, invasion and migration of PC-3 and LNCaP cells. The ‘rescue’ experiments confirmed that miR-145-3p and miR-145-5p directly targeted MTDH to negatively regulate its expression, thereby regulating cell viability and cell invasion and migration. However, this study still had some limitations, as for example, the ‘rescue’ experiments were not validated in DU145 cells, and the sample size of the patients with PCa was not large enough. Thus, we aim to carry out a more comprehensive study on miR-145-5p/miR-145-3p and MTDH in the future.
Collectively, this study, for the first time, at least to the best of our knowledge, demonstrated the effects of the miR-145-5p/MTDH and miR-145-3p/MTDH pathways on the growth and metastasis of PCa cell lines in vitro, and suggests that these pathways may become novel treatment targets for PCa.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors’ contributions
DP made substantial contributions to the conception and design of the study. ZJ, WL and ZD was involved in data acquisition, data analysis and interpretation. DP, WL and ZD drafted the article or critically revised it for important intellectual content. All authors agree to be accountable for all aspects of the study in ensuring that questions related to the accuracy or integrity of the study are appropriately investigated and resolved. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committees of the First Affiliated Hospital and College of Clinical Medicine of Henan University of Science and Technology approved the study. A total of 46 cases of patients signed informed consent forms prior to participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have not competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Cotignola J, Leonardi DB, Shahabi A, Acuña AD, Stern MC, Navone N, Scorticati C, De Siervi A, Mazza O, Vazquez E. Glutathione-S-transferase (GST) polymorphisms are associated with relapse after radical prostatectomy. Prostate Cancer Prostatic Dis. 2013;16:28–34. doi: 10.1038/pcan.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isaacs JT, Schulze H, Coffey DS. Development of androgen resistance in prostatic cancer. Prog Clin Biol Res. 1987;243A:21–31. [PubMed] [Google Scholar]
- 4.Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169:517–523. doi: 10.1016/S0022-5347(05)63946-8. [DOI] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 14.Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- 15.Wach S, Nolte E, Szczyrba J, Stöhr R, Hartmann A, Ørntoft T, Dyrskjøt L, Eltze E, Wieland W, Keck B, et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 16.Szczyrba J, Löprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 18.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szczyrba J, Nolte E, Wach S, Kremmer E, Stöhr R, Hartmann A, Wieland W, Wullich B, Grässer FA. Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol Cancer Res. 2011;9:791–800. doi: 10.1158/1541-7786.MCR-10-0573. [DOI] [PubMed] [Google Scholar]
- 20.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/S1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 21.Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao W, Volsky DJ, Fisher PB. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592–3602. doi: 10.1038/sj.onc.1205445. [DOI] [PubMed] [Google Scholar]
- 22.Li JW, Huang CZ, Li JH, Yuan JH, Chen QH, Zhang WF, Xu ZS, Liu YP, Li Y, Zhan MX, et al. Knockdown of metadherin inhibits cell proliferation and migration in colorectal cancer. Oncol Rep. 2018;40:2215–2223. doi: 10.3892/or.2018.6581. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Li XY, Long M, Wang X, Gao ZW, Cui Y, Ren J, Zhang Z, Liu C, Dong K, et al. The FBXW7 tumor suppressor inhibits breast cancer proliferation and promotes apoptosis by targeting MTDH for degradation. Neoplasma. 2018;65:201–209. doi: 10.4149/neo_2018_170228N149. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Wu H, Wang X, Wang C, Gan L, Zhu J, Tong J, Li Z. Huaier Granule extract inhibit the proliferation and metastasis of lung cancer cells through down-regulation of MTDH, JAK2/STAT3 and MAPK signaling pathways. Biomed Pharmacother. 2018;101:311–321. doi: 10.1016/j.biopha.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Ma Z, Chen Y, Dong S, Xu X, Liu J, Song P, Yu C, Dai L. AEG-1 mRNA expression in non-small cell lung cancer is associated with increased tumor angiogenesis. Pathol Res Pract. 2017;213:1257–1263. doi: 10.1016/j.prp.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Huang LL, Wang Z, Cao CJ, Ke ZF, Wang F, Wang R, Luo CQ, Lu X, Wang LT. AEG-1 associates with metastasis in papillary thyroid cancer through upregulation of MMP2/9. Int J Oncol. 2017;51:812–822. doi: 10.3892/ijo.2017.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Q, Han Y, Zhu D, Li Z, Shan S, Jin W, Lu Q, Ren T. miR-145 and miR-497 suppress TGF-β-induced epithelial-mesenchymal transition of non-small cell lung cancer by targeting MTDH. Cancer Cell Int. 2018;18:105. doi: 10.1186/s12935-018-0601-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z, Ma Y, Pan Y, Zhu H, Yu C, Sun C. MiR-1297 suppresses pancreatic cancer cell proliferation and metastasis by targeting MTDH. Mol Cell Probes. 2018;40:19–26. doi: 10.1016/j.mcp.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–1337. doi: 10.1016/j.biopha.2017.08.143. [DOI] [PubMed] [Google Scholar]
- 30.Qiao W, Cao N, Yang L. MicroRNA-154 inhibits the growth and metastasis of gastric cancer cells by directly targeting MTDH. Oncol Lett. 2017;14:3268–3274. doi: 10.3892/ol.2017.6558. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/S1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 32.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Gu C, Feng L, Peng H, Yang H, Feng Z, Yang Y. MTDH is an oncogene in multiple myeloma, which is suppressed by Bortezomib treatment. Oncotarget. 2016;7:4559–4569. doi: 10.18632/oncotarget.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noch EK, Khalili K. The role of AEG-1/MTDH/LYRIC in the pathogenesis of central nervous system disease. Adv Cancer Res. 2013;120:159–192. doi: 10.1016/B978-0-12-401676-7.00006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS, Li M. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- 37.Liu L, Wu J, Ying Z, Chen B, Han A, Liang Y, Song L, Yuan J, Li J, Li M. Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res. 2010;70:3750–3759. doi: 10.1158/0008-5472.CAN-09-3838. [DOI] [PubMed] [Google Scholar]
- 38.Yu C, Chen K, Zheng H, Guo X, Jia W, Li M, Zeng M, Li J, Song L. Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis. 2009;30:894–901. doi: 10.1093/carcin/bgp064. [DOI] [PubMed] [Google Scholar]
- 39.Song L, Li W, Zhang H, Liao W, Dai T, Yu C, Ding X, Zhang L, Li J. Over-expression of AEG-1 significantly associates with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. J Pathol. 2009;219:317–326. doi: 10.1002/path.2595. [DOI] [PubMed] [Google Scholar]
- 40.Emdad L, Sarkar D, Su ZZ, Randolph A, Boukerche H, Valerie K, Fisher PB. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: Implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 41.Li C, Wu X, Zhang W, Li J, Liu H, Hao M, Wang J, Zhang H, Yang G, Hao M, et al. AEG-1 Promotes Metastasis Through Downstream AKR1C2 and NF1 in Liver Cancer. Oncol Res. 2014;22:203–211. doi: 10.3727/096504015X14386062091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu DC, Yang ZL. MTDH and EphA7 are markers for metastasis and poor prognosis of gallbladder adenocarcinoma. Diagn Cytopathol. 2013;41:199–205. doi: 10.1002/dc.21821. [DOI] [PubMed] [Google Scholar]
- 43.Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 45.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 46.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, Senzer N, Eklund AC, Han J, Nemunaitis J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–216. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Lv L, Li Y, Ji H. MicroRNA 655 suppresses cell proliferation and invasion in oral squamous cell carcinoma by directly targeting metadherin and regulating the PTEN/AKT pathway. Mol Med Rep. 2018;18:3106–3114. doi: 10.3892/mmr.2018.9292. [DOI] [PubMed] [Google Scholar]
- 48.Matsushita R, Yoshino H, Enokida H, Goto Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M, Seki N. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7:28460–28487. doi: 10.18632/oncotarget.8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Misono S, Seki N, Mizuno K, Yamada Y, Uchida A, Arai T, Kumamoto T, Sanada H, Suetsugu T, Inoue H. Dual strands of the miR-145 duplex (miR-145-5p and miR-145-3p) regulate oncogenes in lung adenocarcinoma pathogenesis. J Hum Genet. 2018;63:1015–1028. doi: 10.1038/s10038-018-0497-9. [DOI] [PubMed] [Google Scholar]
- 50.Mataki H, Seki N, Mizuno K, Nohata N, Kamikawaji K, Kumamoto T, Koshizuka K, Goto Y, Inoue H. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget. 2016;7:72084–72098. doi: 10.18632/oncotarget.12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ, et al. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Invest. 2015;33:251–258. doi: 10.3109/07357907.2015.1025407. [DOI] [PubMed] [Google Scholar]
- 52.Goto Y, Kurozumi A, Arai T, Nohata N, Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y, et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br J Cancer. 2017;117:409–420. doi: 10.1038/bjc.2017.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.