Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common type of renal cell carcinoma and the incidence of this disease is increasing. The present study aimed to investigate the role of homeobox A6 (HOXA6) in the proliferation and apoptosis of ccRCC cells. Analysis of the GSE6344 dataset and immunohistochemistry revealed that the mRNA and protein expression levels of HOXA6 were suppressed in ccRCC tissues. To evaluate the roles of HOXA6 in cell proliferation and apoptosis, ccRCC cell lines (786-O and 769-P) were transfected with plasmids expressing HOXA6, empty vector, short hairpin (sh)HOXA6 and non-targeting shRNA (NC). Cell Counting Kit-8, colony formation and 5-ethynyl-2′-deoxyuridine staining assays were performed to analyze cell proliferation. In addition, Caspase-Glo and terminal deoxynucleotidyl transferase dUTP nick end labeling assays were performed to detect apoptosis. Furthermore, the cell cycle and apoptotic rates of 786-O and 769-P cells were analyzed by flow cytometry. The results demonstrated that, compared with the empty vector group, the proliferation of 786-O and 769-P cells decreased following HOXA6 overexpression; however, compared with the NC group, cell proliferation increased in the shHOXA6 group. The rate of apoptosis of HOXA6-overexpressing cells was increased compared with the empty vector group, while the rate of apoptosis in the shHOXA6 group was reduced compared with the NC group. In addition, flow cytometry demonstrated that upregulated HOXA6 expression levels could inhibit the cell cycle at the G0/G1 phase. Western blotting revealed that the expression levels of phosphoinositide 3-kinase (PI3K), phosphorylated (p)-protein kinase B (Akt), mitogen-activated protein kinase kinase, p-extracellular signal-regulated kinase (ERK) and B-cell lymphoma 2 (Bcl-2) were suppressed in cells overexpressing HOXA6; however, the protein expression levels of phosphatase and tensin homolog, Bcl-2-associated X protein, cleaved caspase-3 and cleaved-poly (ADP-ribose) polymerase were increased compared with the empty vector group. Opposing results were reported for the shHOXA6 group compared with the NC group. In summary, the results demonstrated that HOXA6 suppresses cell proliferation and promotes apoptosis, which may occur via inhibition of the PI3K/Akt/ERK cascade. These findings indicate the role of HOXA6 in ccRCC; however, the underlying mechanism requires further investigation.
Keywords: homeobox A6, cell proliferation, apoptosis, clear cell renal cell carcinoma, PI3K, Akt
Introduction
Renal cell carcinoma (RCC) is a common type of cancer worldwide and the incidence of this disease is increasing; the incidence rate of RCC was 10.6/100,000 in 2001, which increased to 12.4/100,000 in 2010 (1,2). Genetic mutations are an important risk factor for the development of RCC (3), which can be detected at early stages. Systemic therapeutic agents, including sunitinib and temsirolimus, have been used to treat RCC (4). The presence of smaller tumors leads to fewer episodes of relapse and improved prognosis (5). Further investigation is required to determine the molecular mechanisms underlying the development of clear cell RCC (ccRCC), the most common type of RCC.
The phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway regulates a series of cellular processes, including the cell cycle, proliferation, apoptosis, autophagy and metastasis in various types of cancer (6). The PI3K/Akt signaling pathway serves its role by regulating numerous effectors, particularly factors of the mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) pathway (7). Due to its key roles in cancer, several potential molecular targets of the PI3K/Akt signaling pathway have been proposed in cancer therapy (8). Furthermore, the PI3K/Akt signaling pathway has been demonstrated to be highly activated and serves a critical role in ccRCC (9,10).
Homeobox A6 (HOXA6) encodes a transcription factor that regulates gene expression and cellular differentiation. HOXA6 has been identified to serve important roles in cell proliferation, differentiation and invasion (11,12). It has been reported that HOXA6 may serve a role in a variety of cancer types; however, the roles of HOXA6 in these cancer types were inconsistent. HOXA6 was observed to be overexpressed in tumor tissues and promoted the progression of cancer (13-15); however, HOXA6 was also determined to be hypermethylated in certain malignancies, indicating reduced expression (16,17). In addition, HOXA6 expression has been reported to be suppressed in breast cancer tissues (18). At present, to the best of our knowledge, extensive research into the roles of HOXA6 in ccRCC has not been conducted. Therefore, the present study aimed to determine the expression profile of HOXA6 in ccRCC. The results revealed that HOXA6 was downregulated in ccRCC; however, the role of HOXA6 in ccRCC and its underlying mechanism requires further investigation. In the present study, the role of HOXA6 in cell proliferation and apoptosis was analyzed, and the association between HOXA6 and the PI3K/Akt signaling pathway in ccRCC was determined.
Materials and methods
Confirmation of HOXA6 expression and survival analysis
Data of HOXA6 mRNA expression was obtained from the GSE6344 dataset (19,20) of the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The tissue samples included in GSE6344 consisted of ten patient-matched normal renal cortex and ccRCC tissues; five samples were of stage I and five were of stage II ccRCC, according to the TNM staging system (21). All patients were diagnosed with localized disease; stage I tumors were <7 cm and stage II tumors were >7 cm. Gene Expression Profiling Interactive Analysis (GEPIA) (22) was performed to determine the association between the mRNA expression levels of HOXA6 and the survival of patients with ccRCC.
Tissue specimens
The present study was approved by the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (Chongqing, China; approval no. 20185501). Written informed consent was obtained from all patients. A total of 25 pairs of ccRCC and adjacent non-tumor tissue specimens were collected at the Department of Urology between June 2018 and August 2018. The distance between the ccRCC sample and the non-tumor tissue sample was ≥2 cm. Among the 25 patients, 14 patients underwent radical nephrectomy and 11 underwent partial nephrectomy without any pre-operative treatment. The median age of the patients was 53 years (range, 42-78 years). The clinicopathological features of the patients are presented in Table I. Pathological diagnosis of the specimens was based on the pathological report of the Department of Pathology of Chongqing Medical University.
Table I.
Clinicopathological characteristics of patients with ccRCC.
| Case no. | Age, years | Sex | Diagnosis | TNM stage | Tumor size, cm |
|---|---|---|---|---|---|
| 1 | 52 | Male | Right renal ccRCC | T1aN0M0 | 2.7 |
| 2 | 66 | Male | Left renal ccRCC | T2aN0M0 | 7.2 |
| 3 | 63 | Male | Right renal ccRCC | T1aN0M0 | 2.4 |
| 4 | 60 | Male | Left renal ccRCC | T1aN0M0 | 3.1 |
| 5 | 66 | Female | Right renal ccRCC | T1aN0M0 | 2.6 |
| 6 | 74 | Male | Left renal ccRCC | T1aN0M0 | 2.5 |
| 7 | 47 | Female | Left renal ccRCC | T1aN0M0 | 3.2 |
| 8 | 68 | Male | Right renal ccRCC | T1aN0M0 | 2.7 |
| 9 | 78 | Male | Right renal ccRCC | T1aN0M0 | 2.0 |
| 10 | 67 | Male | Right renal ccRCC | T2aN0M0 | 7.1 |
| 11 | 72 | Female | Left renal ccRCC | T1aN0M0 | 2.1 |
| 12 | 49 | Male | Right renal ccRCC | T1aN0M0 | 2.5 |
| 13 | 47 | Male | Left renal ccRCC | T2aN0M0 | 8.6 |
| 14 | 44 | Male | Left renal ccRCC | T1bN0M0 | 4.7 |
| 15 | 53 | Female | Left renal ccRCC | T3aN0M0 | 4.0 |
| 16 | 64 | Male | Left renal ccRCC | T3aN0M0 | 5.0 |
| 17 | 50 | Male | Left renal ccRCC | T1bN0M0 | 5.0 |
| 18 | 47 | Male | Right renal ccRCC | T1bN0M0 | 7.0 |
| 19 | 50 | Female | Left renal ccRCC | T1aN0M0 | 3.8 |
| 20 | 50 | Female | Left renal ccRCC | T1bN0M0 | 4.5 |
| 21 | 62 | Male | Left renal ccRCC | T1bN0M0 | 6.5 |
| 22 | 43 | Male | Left renal ccRCC | T1aN0M0 | 2.5 |
| 23 | 42 | Male | Left renal ccRCC | T1aN0M0 | 3.0 |
| 24 | 53 | Male | Left renal ccRCC | T1bN0M0 | 5.0 |
| 25 | 66 | Female | Right renal ccRCC | T3aN0M0 | 3.5 |
ccRCC, clear cell renal cell carcinoma.
Immunohistochemistry (IHC)
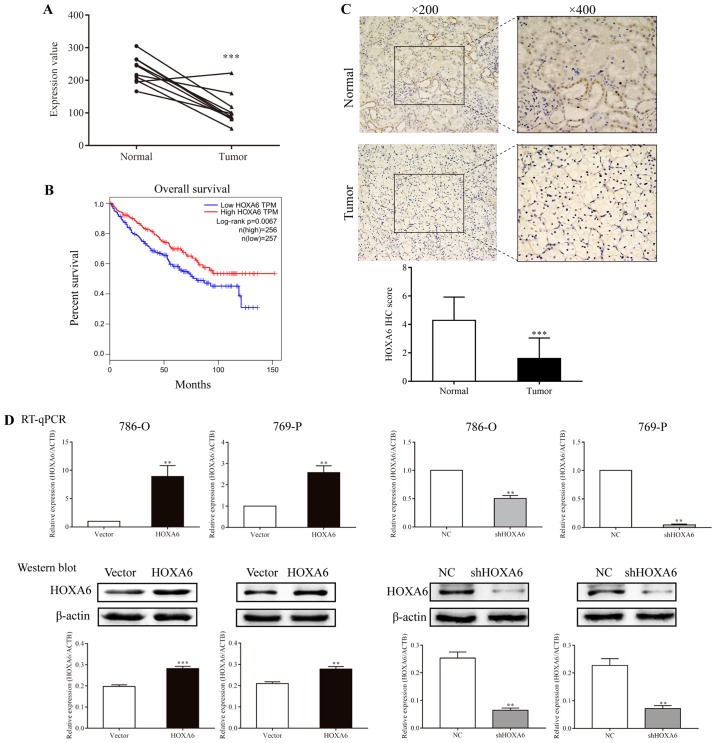
Tissues were fixed with 10% formalin at room temperature for 4-6 h. Subsequently, paraffin-embedded tissue sections (5-µm) were dewaxed and rehydrated in 100, 95, 85, 75 and 50% ethanol, boiled at 100°C in 0.01 mol/l sodium citrate buffer (pH 6.0) for 15 min for antigen retrieval and incubated with 3% hydrogen peroxide for 20 min to inhibit endogenous peroxidase activity. Samples were blocked with normal goat serum (reagent B in the SP-9001 kit; OriGene Technologies, Inc.) at 37°C for 30 min and incubated with rabbit anti-HOXA6 (cat. no. bs-11294R, 1:400; BIOSS) overnight at 4°C. A horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG working solution (reagent C in the SP-9001 kit; OriGene Technologies, Inc.) was then applied for 30 min at 37°C. Subsequently, sections were stained at room temperature with 3,3′-diaminobenzidene (OriGene Technologies, Inc.) for 10 sec and hematoxylin (OriGene Technologies, Inc.) for 30 sec at room temperature. Negative control specimens were incubated without anti-HOXA6.
The score criterion was set according to the intensity and the extent of section staining (percentage of positive cells). The intensity scores were as follows: 0, no staining; 1, weak; 2, moderate; and 3, intense. The scores for the extent of staining were as follows: 0, 0%; 1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, 76-100%. The IHC scores were calculated by multiplying the intensity scores with the extent of staining scores (0-12). IHC scores <3 were defined as low expression, while scores ≥4 were defined as high expression. A light microscope was used to capture the images (magnification, ×200 and ×400). The ccRCC subtype and IHC scores were confirmed independently by two pathologists.
Cell culture and transfection
The human ccRCC cell lines, 786-O and 769-P, were purchased from the American Type Culture Collection and cultured in RPMI-1640 medium (HyClone; GE Healthcare) containing 10% fetal bovine serum (PAN-Biotech) at 37°C in 5% CO2.
The plasmids were supplied by GeneCopoeia, Inc., including HOXA6-expressing plasmid, empty vector (termed vector; pReceiver-M03), and plasmids containing short hairpin (sh)HOXA6 (5′-CCTTGTTTCTACCAACAGTCC-3′) and non-targeting shRNA (NC; 5′-GCTTCGCGCCGTAGT CTTA-3′). 786-O and 769-P cells were transfected using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer’s protocol. Cells were seeded into a 6-well plate at a density of ~2×105 cells per well, and a total of 4 µg plasmids were added for transfection at 50-75% confluence.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
At 48 h post-transfection, total RNA was extracted from cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and reverse transcribed with the GoScript™ Reverse Transcription system (Promega Corporation) at 25°C for 5 min, 42°C for 60 min, 70°C for 15 min and 4°C until the subsequent step. The primers used for qPCR were as follows (15): HOXA6 forward, 5′-TACACGCGCTACCAGA CAC-3′ and reverse, 5′-GCGTGGAATTGATGAGCTTG TTT-3′, and β-actin forward, 5′-TGGAACGGTGAAGGTGA CAG-3′ and reverse, 5′-AACAACGCATCTCATATTTG GAA-3′. qPCR was performed using the GoTaq qPCR Master mix (Promega Corporation), with the following thermocycling conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min, followed by 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec, according to the manufacturer’s protocol. Data were analyzed using the 2−∆∆Cq method (23).
Western blotting
At 72 h post-transfection, total proteins were extracted from cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) and the protein concentration was measured via the bicinchoninic acid method. Proteins (40 µg) were separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Subsequently, the PVDF membrane was blocked with 5% skim milk at room temperature for 2 h and incubated with primary antibodies at 4°C overnight. The membrane was then incubated with an HRP-conjugated secondary antibody (1:3,000, cat. no. bs-0295G; BIOSS) at 37°C for 2 h. Bands were visualized using an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology). The grayscale value of the images was analyzed using Fusion FX Spectra software (version 7; Vilber). The primary antibodies were as follows: Anti-HOXA6 (1:800; cat. no. bs-11294R; BIOSS), anti-PI3K (1:1,000; cat. no. 4257; Cell Signaling Technology, Inc.), anti-Akt (1:1,000; cat. no. 4691; Cell Signaling Technology, Inc.), anti-phosphorylated (p)-Akt (1:1,000; cat. no. 9271; Cell Signaling Technology, Inc.), anti-PTEN (1:1,000; cat. no. 9188; Cell Signaling Technology, Inc.), anti-MEK (1:1,000; cat. no. 8727; Cell Signaling Technology, Inc.), anti-ERK (1:1,000; cat. no. 4695; Cell Signaling Technology, Inc.), anti-p-ERK (1:1,000; cat. no. 4370; Cell Signaling Technology, Inc.), anti-Bax (1:1,000; cat. no. ab32503; Abcam), anti-Bcl-2 (1:1,000; cat. no. ab32124; Abcam), anti-cleaved-PARP (1:1,000; cat. no. ab32561; Abcam), anti-cleaved-caspase3 (1:1,000; cat. no. ab2302; Abcam), anti-caspase3 (1:1,000; cat. no. ab44976; Abcam), anti-β-actin (1:1,000; cat. no. ab8227; Abcam).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 regent (Dojindo Molecular Technologies, Inc.) was used to detect cell proliferation (24). At 48 h post-transfection, cells were seeded in a 96-well plate (4×103 cells/well). At each time point for detection (24, 48 and 72 h), cells were incubated with CCK-8 reagent at 37°C for 2 h and the absorbance at 450 nm was measured.
Colony formation assay
At 48 h post-transfection, cells were seeded in a 6-well plate (500 cells/well), cultured at 37°C in 5% CO2 for 2 weeks, then fixed using 4% paraformaldehyde at room temperature for 15 min and stained with 0.5% gentian violet (Beyotime Institute of Biotechnology) at room temperature for 15 min. Clones consisting of ≥50 cells were counted under a light microscope (magnification, ×40) (25,26).
5-Ethynyl-2′-deoxyuridine (EdU) staining
Cell proliferation was also analyzed using a Cell-Light EdU Apollo643 in vitro kit (cat. no. C10310-2; Guangzhou RiboBio Co., Ltd.), according to the manufacturer’s protocol (27,28). A fluorescence microscope (Olympus Corporation) was used for analysis and images were obtained (magnification, ×100).
Caspase-Glo assay
The activities of caspases-3, -7, -8 and -9 were measured using Caspase-Glo 3/7, 8 and 9 kits (cat. nos. G8090, G8200 and G8210, respectively; Promega Corporation), according to the manufacturer’s protocol. Transfected cells were seeded (5×103 cells/well) into a 96-well plate and incubated with Caspase-Glo reagents for 30 min. The luminescence was then detected with a microplate reader.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)
Transfected cells were seeded onto a climbing slide (5×103 cells/well), cultured overnight and stained with a TUNEL reagent kit (In Situ Cell Death Detection kit, POD; cat. no. 11684817910; Roche Diagnostics) (29) at 37°C for 1-2 h and DAPI at 37°C for 5 min. Images were obtained using a fluorescence microscope (magnification, ×100; Olympus Corporation).
Flow cytometry
To analyze the cell cycle, cells were suspended in PBS buffer at 48 h post-transfection, fixed in 75% ethanol at 4°C overnight and stained with propidium iodide (PI) at room temperature for 30 min. For the analysis of cell apoptosis, harvested cells were suspended in PBS and stained using an Annexin V-fluorescein isothiocyanate/PI kit (cat. no. 556570, FITC Annexin V Apoptosis Detection kit II, BD Biosciences). Following staining, cells were immediately detected using a flow cytometer (FACSCalibur; BD Biosciences), and analyzed with ModFit LT™ V3.3 (Verity Software House, Inc.) for cell cycle analysis and FlowJo 10 (FlowJo LLC) for apoptosis analysis (30,31).
Cell migration
Cell migration was evaluated using Transwell and wound-healing assays with the 786-O cell line. For the Transwell assay, 5×104 cells suspended in 200 µl RPMI-1640 medium was added to the upper chamber and 700 µl complete medium was added to the lower chamber. After incubation for 48 h, the migrated cells were fixed. The fixed cells were then stained with crystal violet at room temperature for 15 min. The stained cells were viewed and counted in five random fields of view under an inverted phase contrast microscope (magnification, ×200). For the wound-healing assay, the transfected cells were seeded in a 6-well plate (5×105 cells/well). At confluence, the cell monolayer was scraped off and maintained in RPMI-1640. After 0, 24 and 48 h of incubation, an inverted microscope (magnification, ×100) was used to capture the images.
Statistical analysis
Data were analyzed using SPSS 22.0 (IBM Corp.) and three independent experiments were performed. The data are presented as the mean ± standard deviation. CCK-8 and wound-healing results were statistically analyzed using two-way ANOVA. All other data were compared using unpaired Student’s t-test. P<0.05 was considered to indicate a statistically significant difference.
Results
HOXA6 is downregulated in ccRCC specimens
The GSE6344 database contains ten paired ccRCC and normal tissues. The mRNA expression levels of HOXA6 were extracted from this database and analyzed. The results revealed that HOXA6 mRNA expression was significantly increased in normal tissues compared with in carcinoma tissues (Fig. 1A). Subsequently, GEPIA online analysis demonstrated that patients with lower expression levels of HOXA6 exhibited significantly poorer survival (Fig. 1B). Additionally, a total of 25 paired ccRCC and adjacent non-tumor specimens were analyzed by IHC. The majority of normal specimens (15/25) exhibited high expression levels of HOXA6, whereas the majority of tumor specimens (23/25) demonstrated low HOXA6 protein expression levels. HOXA6 was notably expressed in the nucleus of renal tubular epithelial cells. Furthermore, the protein expression levels of HOXA6 in normal tissues were significantly increased compared with in paired carcinoma tissues (Fig. 1C). These results suggest that HOXA6 protein expression is downregulated in ccRCC, which is in accordance with the findings of GSE6344 analysis.
Figure 1.
Expression of HOXA6 in carcinoma tissues and transfected cell lines, and survival analysis. (A) The mRNA expression levels of HOXA6 in ten paired ccRCC tumor and normal tissues extracted from GSE6344. ***P<0.001 vs. Normal. (B) Survival analysis of patients with ccRCC, which was performed using Gene Expression Profiling Interactive Analysis. (C) The expression of HOXA6 protein in ccRCC and normal tissues as detected by immunohistochemistry. ***P<0.001 vs. Normal. (D) Expression of HOXA6 in transfected cell lines. **P<0.01 vs. vector or NC. ***P<0.001 vs. vector. ccRCC, clear cell renal cell carcinoma; HOXA6, homeobox A6; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; vector, vector-only control; NC, negative control; shHOXA6, HOXA6 short hairpin RNA; ACTB, β-actin; TPM, transcripts per kilobase of exon model per million mapped reads.
Detection of HOXA6 expression in transfected cells
786-O and 769-P cells were transfected with HOXA6 plasmid, empty vector, NC or shHOXA6, and HOXA6 expression was determined by RT-qPCR and western blotting. The results revealed that the expression of HOXA6 in 786-O and 769-P cells overexpressing HOXA6 was increased compared with the vector groups (RT-qPCR, P=0.0032 and P=0.0058; western blotting, P<0.001 and P=0.0020, respectively). Compared with the corresponding NC groups of 786-O and 769-P cells, HOXA6 expression in the shHOXA6 groups was significantly lower (RT-qPCR, P=0.0036 and P=0.0050; western blotting, P=0.0019 and P=0.0030, respectively; Fig. 1D). These results indicated that HOXA6 expression was successfully promoted and inhibited in the respective groups of transfected cells.
HOXA6 inhibits the proliferation of 786-O and 769-P cells
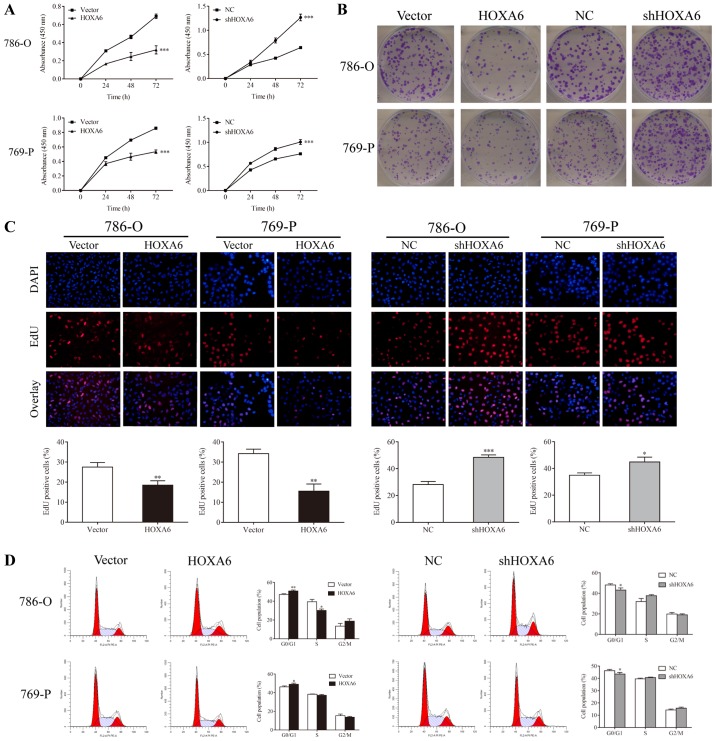
CCK-8, colony formation, EdU and flow cytometry assays were performed to determine the proliferation of transfected 786-O and 769-P cells. The CCK-8 assay revealed that HOXA6-overexpressing cells exhibited reduced proliferation compared with the vector groups (P<0.001), whereas the proliferation of cells expressing shHOXA6 significantly increased compared with the NC groups (P<0.001; Fig. 2A). In addition, fewer colonies in the HOXA6 overexpression groups (786-O, 95.67±11.71; 769-P, 143.33±10.02) were observed compared with the vector groups (786-O, 199.33±23.25, P=0.0066; 769-P, 189.33±17.04, P=0.0238); however, the number of colonies was significantly increased in the shHOXA6 groups (786-O, 306.00±22.72; 769-P, 338.33±28.04) compared with the corresponding NC groups (786-O, 196.33±17.67, P=0.0034; 769-P, 213.00±13.11, P=0.0071; Fig. 2B). Additionally, EdU staining revealed that, compared with the vector groups (786-O, 27.53±2.27%; 769-P, 34.24±1.28%), the proliferation of HOXA6-overexpressing cells (786-O, 18.44±2.26%, P=0.0079; 769-P, 15.52±2.07%, P=0.0015) was significantly inhibited. By contrast, the proliferation of the shHOXA6 groups (786-O, 48.41±1.94%; 769-P, 44.86±3.60%) was promoted compared with the corresponding NC groups (786-O, 28.19±2.28%, P<0.001; 769-P, 34.99±1.68%, P=0.0141; Fig. 2C). Furthermore, the cell cycle of transfected cells was analyzed by PI staining and flow cytometry. Compared with the vector groups, the number of G0/G1 phase cells was significantly increased in the HOXA6 overexpression groups (786-O, P=0.0072; 769-P, P=0.0380). By contrast, the number of G0/G1 phase cells was significantly decreased in the shHOXA6 expression groups compared with the corresponding NC groups (786-O, P=0.0332; 769-P, P=0.0460; Fig. 2D). These results indicate that HOXA6 suppresses cell proliferation by inhibiting the cell cycle at the G0/G1 phase in ccRCC.
Figure 2.
HOXA6 serves a role in the proliferation of cells. (A) Cell proliferation was determined by a Cell Counting Kit-8 assay. ***P<0.001 vs. vector or NC. (B) Cell proliferation was detected by a colony formation assay. (C) Cells stained with DAPI and EdU were considered as positive for proliferation. **P<0.01 vs. vector; *P<0.05, ***P<0.001 vs. NC (magnification, ×200). (D) Cell cycle analysis via flow cytometry. *P<0.05, **P<0.01 vs. vector or NC. EdU, 5-ethynyl-2′-deoxyuridine; HOXA6, homeobox A6; vector, vector-only control; NC, negative control; shHOXA6, HOXA6 short hairpin RNA.
HOXA6 induces the apoptosis of 786-O and 769-P cells
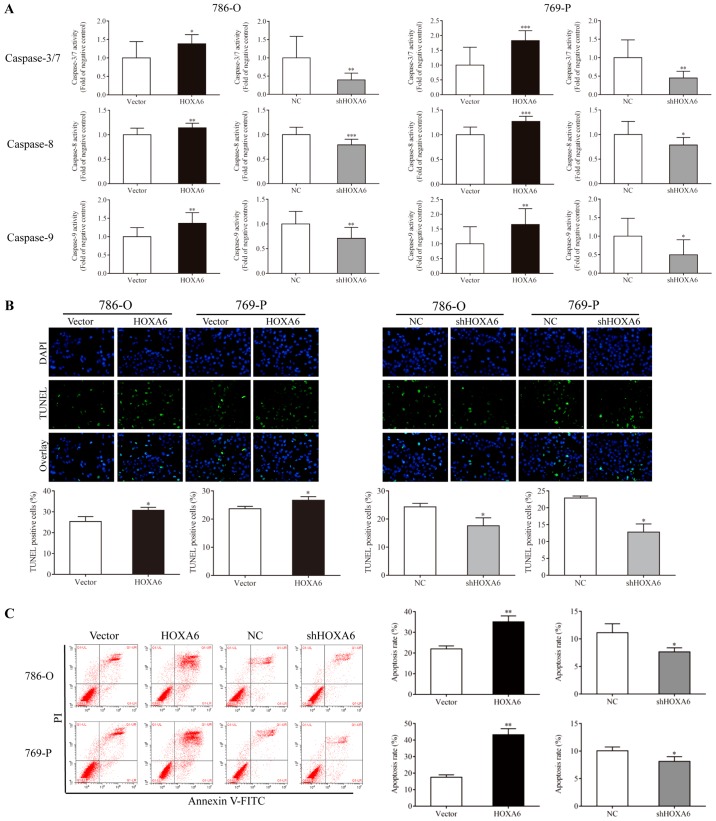
Caspase-Glo, TUNEL and flow cytometry assays were performed to analyze the apoptosis of transfected cells. A Caspase-Glo assay was performed to detect the activities of caspases-3/7, -8 and -9. Compared with the vector group, the activities of caspases-3/7, -8 and -9 (were significantly increased in the HOXA6-overexpressing 786-O and 769-P cell groups. By contrast, the activities of caspases-3/7 were significantly reduced in the shHOXA6-expressing 786-O and 769-P cell groups compared with the corresponding NC groups (Fig. 3A). The TUNEL assay also indicated that the apoptotic rates of the HOXA6 overexpression groups (786-O, 30.80±1.34%; 769-P; 26.72±1.28%) were significantly increased compared with the vector groups (786-O, 25.31±2.41%, P=0.0384; 769-P, 23.69±0.86%, P=0.0332). However, the apoptotic rates were significantly decreased in the shHOXA6 groups (786-O, 17.61±2.82%; 769-P, 12.83±2.39%) compared with the corresponding NC groups (786-O, 24.31±1.30%, P=0.0372; 769-P, 22.92±0.58%, P=0.0141; Fig. 3B). In addition, flow cytometry was performed to analyze the apoptosis of transfected cells. The results revealed that, compared with the vector group, upregulation of HOXA6 significantly induced cell apoptosis (786-O, P=0.0067; 769-P, P=0.0027), while downregulation of HOXA6 suppressed apoptosis compared with the corresponding NC groups (786-O, P=0.0475; 769-P, P=0.0418; Fig. 3C). These results demonstrated that HOXA6 could induce the apoptosis of ccRCC cells.
Figure 3.
Cell apoptosis as analyzed via Caspase-Glo, TUNEL and flow cytometry assays. (A) Caspase-Glo analysis was performed to detect the apoptosis of cells. *P<0.05, **P<0.01, ***P<0.001 vs. vector or NC. (B) Cells stained with TUNEL and DAPI were considered as apoptotic, *P<0.05 vs. vector or NC (magnification, ×200). (C) Cell apoptosis as analyzed by flow cytometry. *P<0.05 vs. NC, **P<0.01 vs. vector .TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; HOXA6, homeobox A6; vector, vector-only control; NC, negative control; shHOXA6, HOXA6 short hairpin RNA; PI, propidium iodide; FITC, fluorescein isothiocyanate.
HOXA6 inhibits the PI3K/Akt signaling pathway
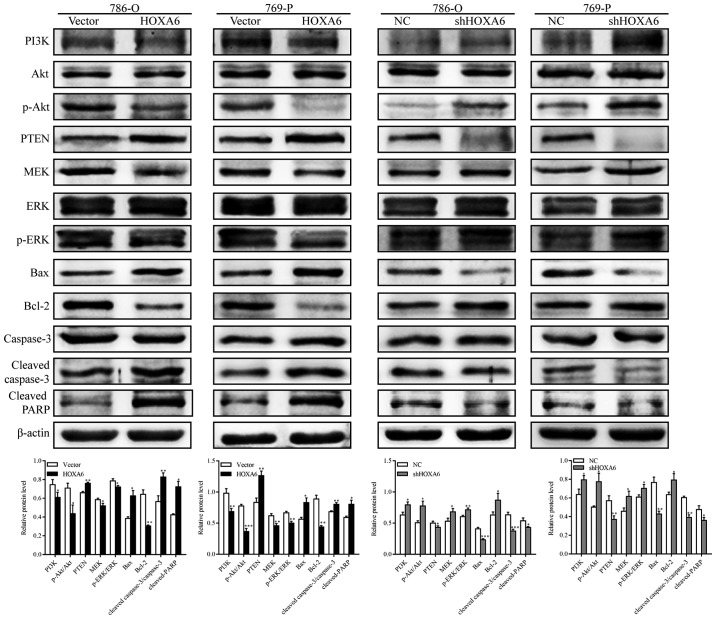
The expression levels of PI3K, Akt, p-Akt, phosphatase and tensin homolog (PTEN), MEK, ERK, p-ERK, B-cell lymphoma 2 (Bcl-2)-associated X protein (Bax), Bcl-2, caspase-3, cleaved caspase-3 and cleaved poly (ADP-ribose) polymerase (PARP) were analyzed by western blotting. The expression levels of PI3K, p-Akt, MEK, p-ERK and Bcl-2 were significantly suppressed in the HOXA6 overexpression groups, but were significantly upregulated following downregulation of HOXA6. The expression levels of PTEN, Bax, cleaved caspase-3 and cleaved PARP were significantly increased following HOXA6 overexpression compared with the vector group, but were significantly decreased in the shHOXA6 groups compared with the corresponding NC group (Fig. 4). These results suggest that HOXA6 inhibits cell proliferation and induces apoptosis via suppression of the PI3K/Akt signaling pathway.
Figure 4.
Western blotting for the analysis of proteins involved in the PI3K/Akt signaling pathway. Compared with the vector group, the expression levels of PI3K, p-Akt, MEK, p-ERK and Bcl-2 were suppressed in the HOXA6 group, but the expression levels of PTEN, Bax, cleaved caspase-3 and cleaved PARP were increased. Opposing results were observed in the shHOXA6 group compared with the NC group. *P<0.05, **P<0.01, ***P<0.001 vs. vector or NC. Akt, protein kinase B; PI3K, phosphoinositide 3-kinase; p, phosphorylated; PTEN, phosphatase and tensin homolog; MEK, mitogen-activated protein kinase kinase; ERK, extracellular signal-regulated kinase; Bax, B-cell lymphoma 2-associated X protein; Bcl-2, B-cell lymphoma 2; PARP, poly (ADP-ribose) polymerase; HOXA6, homeobox A6; vector, vector-only control; NC, negative control; shHOXA6, HOXA6 short hairpin RNA.
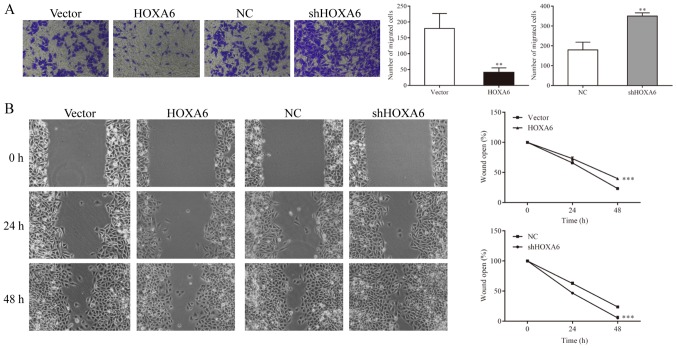
HOXA6 inhibits the migration of 786-O cells
Cell migration was detected by Transwell and wound-healing assays. The Transwell assay results indicated that significantly fewer migrated cells were observed in the HOXA6 overexpression group compared with the vector group (P=0.0082). In addition, the number of migrated cells was significantly increased in the shHOXA6 group compared with the NC group (P=0.0023; Fig.5A). The wound-healing assay results revealed that compared with the vector group, the migration ability was significantly reduced in the HOXA6-overexpressing cells (P<0.001). By contrast, the migration ability was significantly increased in the shHOXA6-expressing cells compared with the NC group (P<0.001; Fig. 5B).
Figure 5.
Cell migration measured by Transwell and wound-healing assays. (A) Transwell assay was performed to evaluate the migration of cells. **P<0.01 vs. vector or NC. (B) Wound-healing assay was performed to detect the migration of cells. ***P<0.001 vs. vector or NC. HOXA6, homeobox A6; vector, vector-only control; NC, negative control; shHOXA6, HOXA6 short hairpin RNA.
Discussion
It has been reported that HOXA6 is upregulated in certain types of cancer and exhibits a role as an oncogene (13-15); however, studies have demonstrated that the CpG islands of HOXA6 are hypermethylated in malignant tumors (16,17). In addition, downregulated HOXA6 expression has been reported in breast cancer tissues (18); therefore, conflicting observations have been reported. In the present study, the expression of HOXA6 in ccRCC was downregulated. To further investigate this finding, the expression of HOXA6 in ccRCC microarrays from a GEO database was analyzed. The expression of HOXA6 mRNA was downregulated in cancer tissues in the GSE6344 dataset. To further verify these results, HOXA6 protein expression in ccRCC tissues was determined by IHC. The results revealed reduced expression of HOXA6 in cancer tissues compared with normal tissues. Furthermore, survival analysis was performed via GEPIA, which was based on data of The Cancer Genome Atlas. This analysis demonstrated that low expression of HOXA6 was associated with poor survival. The aforementioned results indicate that HOXA6 may serve a protective role in ccRCC. Furthermore, following the promotion or suppression of HOXA6 expression in ccRCC cells, alterations in cell proliferation and apoptosis were analyzed in the present study. CCK-8, colony formation, EdU staining and flow cytometry assays indicated that HOXA6 may suppress cell proliferation and inhibit the cell cycle at the G0/G1 phase. Caspase-Glo, TUNEL and flow cytometry assays further revealed that HOXA6 may promote cell apoptosis, and the activities of caspases-3/7, -8 and -9. However, a limitation of the present study is that a rescue experiment of apoptosis by HOXA6 was not performed.
The PI3K/Akt signaling pathway serves an important role in ccRCC for the regulation of cell proliferation, differentiation and survival. PTEN is a crucial inhibitor of the PI3K/Akt signaling pathway and functions as a tumor suppressor (32-34). In the present study, overexpression of HOXA6 promoted the expression of PTEN, leading to reductions in PI3K expression and Akt phosphorylation. p-Akt has a series of downstream effectors and is involved in various sub-pathways, including the mammalian target of rapamycin, glycogen synthase kinase 3, Bcl-2-associated death promoter, ERK, nuclear factor-κB and the c-Jun N-terminal kinase signaling pathways (35). The progression of the cell cycle from the G1 to S-phase is dependent on the activation of ERK (36). MEK is the upstream regulator of ERK, and can induce proliferation and inhibit apoptosis; thus, MEK may be a potential target of cancer therapy (37,38). The present study revealed that upregulation of HOXA6 led to reductions in the levels of p-Akt, increased the expression of MEK and suppressed the phosphorylation of ERK. The ERK signaling pathway has been reported to induce the anti-apoptotic properties of Bcl-2 (39). This supports the results of the present study as the expression of Bcl-2 was suppressed but that of Bax was increased, which leads to a decrease of the Bcl-2/Bax ratio following the upregulation of HOXA6. A decrease in the Bcl-2/Bax ratio has been negatively associated with cell apoptosis (40,41). Caspase-3 is the most important executioner of apoptosis, while PARP is an indicator of caspase-3 activation (42-44). The present study revealed that the protein expression levels of caspase-3 and PARP were upregulated in the HOXA6 group. These findings demonstrated that HOXA6 was involved in regulating cell proliferation and apoptosis, which may occur via the PI3K/Akt/ERK cascade in ccRCC cells.
In the present study, the expression of HOXA6 was suppressed in ccRCC tissues, which was associated with poor survival. In addition, it was identified that HOXA6 may be involved in regulating cell proliferation and apoptosis in ccRCC. Analysis of the pathways associated with these processes revealed that HOXA6 may be involved in the PI3K/Akt signaling pathway; however, the mechanism of how it regulates this pathway requires further investigation. The underlying mechanism of HOXA6 downregulation in ccRCC remains unknown; however, HOXA6 has been reported to be hypermethylated in meningiomas (17). Thus, further investigation is required to determine whether HOXA6 is hypermethylated in ccRCC and the additional roles of HOXA6 in this disease.
In summary, the present study reported that the expression of HOXA6 was downregulated in ccRCC and was negatively associated with the survival of patients. HOXA6 expression was determined to be associated with reduced cell proliferation and increased apoptosis, which may occur via inhibition of the PI3K/Akt/ERK pathway. These results indicate that HOXA6 may serve a role in the regulation of ccRCC; however, the underlying mechanism requires further investigation.
Acknowledgments
The authors would like to thank Dr Lixue Chen (The First Affiliated Hospital of Chongqing Medical University, Chongqing, China) for providing technical support.
Abbreviations
- ccRCC
clear cell renal cell carcinoma
- IHC
immunohistochemistry
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- CCK-8
Cell Counting Kit-8
- EdU
5-ethynyl-2′-deoxyuridine
- TUNEL
terminal deoxynucleotidyl trans-ferase dUTP nick end labeling
Funding
The present study was supported by a grant from the Foundation of Chongqing Science and Technology Commission (grant no. cstc2015shmszx0466).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
FW made substantial contributions to the study conception, design, acquisition of data, analysis and interpretation of the data, and was involved in drafting the manuscript. SW and HT took part in the acquisition, analysis and interpretation of the data. WH and XG were involved in study conception, study design and revising the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was signed by all patients and ethics approval was obtained from the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (Chongqing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.King SC, Pollack LA, Li J, King JB, Master VA. Continued increase in incidence of renal cell carcinoma especially in young patients and high grade disease: United States 2001 to 2010. J Urol. 2014;191:1665–1670. doi: 10.1016/j.juro.2013.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipworth L, Tarone RE, McLaughlin JK. The epidemiology of renal cell carcinoma. J Urol. 2006;176:2353–2358. doi: 10.1016/j.juro.2006.07.130. [DOI] [PubMed] [Google Scholar]
- 4.Banegas MP, Harlan LC, Mann B, Yabroff KR. Renal cell cancer: A shift in approaches for treatment of advanced disease in the United States. J Natl Compr Canc Netw. 2014;12:1271–1279. doi: 10.6004/jnccn.2014.0124. [DOI] [PubMed] [Google Scholar]
- 5.Sunela KL, Lehtinen ET, Kataja MJ, Kujala PM, Soimakallio S, Kellokumpu-Lehtinen PL. Development of renal cell carcinoma (RCC) diagnostics and impact on prognosis. BJU Int. 2014;113:228–235. doi: 10.1111/bju.12242. [DOI] [PubMed] [Google Scholar]
- 6.Roy B, Pattanaik AK, Das J, Bhutia SK, Behera B, Singh P, Maiti TK. Role of PI3K/Akt/mTOR and MEK/ERK pathway in Concanavalin A induced autophagy in HeLa cells. Chem Biol Interact. 2014;210:96–102. doi: 10.1016/j.cbi.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol Biosyst. 2015;11:1946–1954. doi: 10.1039/C5MB00101C. [DOI] [PubMed] [Google Scholar]
- 8.Polivka J, Jr, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Guo H, German P, Bai S, Barnes S, Guo W, Qi X, Lou H, Liang J, Jonasch E, Mills GB, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42:343–353. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F, Wu S, Gou X. Identification of biomarkers and potential molecular mechanisms of clear cell renal cell carcinoma. Neoplasma. 2018;65:242–252. doi: 10.4149/neo_2018_170511N342. [DOI] [PubMed] [Google Scholar]
- 11.Dickson GJ, Kwasniewska A, Mills KI, Lappin TR, Thompson A. Hoxa6 potentiates short-term hemopoietic cell proliferation and extended self-renewal. Exp Hematol. 2009;37:322–333. e323. doi: 10.1016/j.exphem.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Guo YB, Shao YM, Chen J, Xu SB, Zhang XD, Wang MR, Liu HY. Effect of overexpression of HOX genes on its invasive tendency in cerebral glioma. Oncol Lett. 2016;11:75–80. doi: 10.3892/ol.2015.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Liu Y, Yan L, Zhang M, Yu X, Du W, Wang S, Li Q, Chen H, Zhang Y, et al. Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell Death Dis. 2018;9:707. doi: 10.1038/s41419-018-0725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, Wu F, Jiang Z. Effect of HOXA6 on the proliferation, apoptosis, migration and invasion of colorectal cancer cells. Int J Oncol. 2018;52:2093–2100. doi: 10.3892/ijo.2018.4352. [DOI] [PubMed] [Google Scholar]
- 15.Eoh KJ, Kim HJ, Lee J-Y, Nam EJ, Kim S, Kim SW, Kim YT. Upregulation of homeobox gene is correlated with poor survival outcomes in cervical cancer. Oncotarget. 2017;8:84396–84402. doi: 10.18632/oncotarget.21041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xavier FC, Destro MF, Duarte CM, Nunes FD. Epigenetic repression of HOXB cluster in oral cancer cell lines. Arch Oral Biol. 2014;59:783–789. doi: 10.1016/j.archoralbio.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Galani V, Lampri E, Varouktsi A, Alexiou G, Mitselou A, Kyritsis AP. Genetic and epigenetic alterations in meningiomas. Clin Neurol Neurosurg. 2017;158:119–125. doi: 10.1016/j.clineuro.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Hur H, Lee JY, Yun HJ, Park BW, Kim MH. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56:64–71. doi: 10.1007/s12033-013-9682-4. [DOI] [PubMed] [Google Scholar]
- 19.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 20.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, Luxon BA, Sinha M, Anastasiadis PZ, Copland JA. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th edition. Springer; New York, NY: 2010. pp. 547–560. [Google Scholar]
- 22.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Ishiyama M, Miyazono Y, Sasamoto K, Ohkura Y, Ueno K. A highly water-soluble disulfonated tetrazolium salt as a chro-mogenic indicator for NADH as well as cell viability. Talanta. 1997;44:1299–1305. doi: 10.1016/S0039-9140(97)00017-9. [DOI] [PubMed] [Google Scholar]
- 25.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 26.Rafehi H, Orlowski C, Georgiadis GT, Ververis K, El-Osta A, Karagiannis TC. Clonogenic assay: Adherent cells. J Vis Exp. 2011;(49):2573. doi: 10.3791/2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierzyńska-Mach A, Szczurek A, Cella Zanacchi F, Pennacchietti F, Drukała J, Diaspro A, Cremer C, Darzynkiewicz Z, Dobrucki JW. Subnuclear localization, rates and effectiveness of UVC-induced unscheduled DNA synthesis visualized by fluorescence widefield, confocal and super-resolution microscopy. Cell Cycle. 2016;15:1156–1167. doi: 10.1080/15384101.2016.1158377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szeberenyl J. Analysis of the cell cycle by flow cytometry. Biochem Mol Biol Educ. 2007;35:153–154. doi: 10.1002/bmb.41. [DOI] [PubMed] [Google Scholar]
- 31.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 32.Worby CA, Dixon JE. Pten Annu Rev Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 33.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: The long and the short of it. Trends Biochem Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyatt LA, Filbin MT, Keirstead HS. PTEN inhibition enhances neurite outgrowth in human embryonic stem cell-derived neuronal progenitor cells. J Comp Neurol. 2014;522:2741–2755. doi: 10.1002/cne.23580. [DOI] [PubMed] [Google Scholar]
- 35.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nat Rev Clin Oncol. 2014;11:385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 38.Neuzillet C, Tijeras-Raballand A, de Mestier L, Cros J, Faivre S, Raymond E. MEK in cancer and cancer therapy. Pharmacol Ther. 2014;141:160–171. doi: 10.1016/j.pharmthera.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Yang T, Xu F, Sheng Y, Zhang W, Chen Y. A targeted proteomics approach to the quantitative analysis of ERK/Bcl-2-mediated anti-apoptosis and multi-drug resistance in breast cancer. Anal Bioanal Chem. 2016;408:7491–7503. doi: 10.1007/s00216-016-9847-7. [DOI] [PubMed] [Google Scholar]
- 40.Zeren T, Inan S, Vatansever HS, Sayhan S. Significance of apoptosis related proteins on malignant transformation of ovarian tumors: A comparison between Bcl-2/Bax ratio and p53 immunoreactivity. Acta Histochem. 2014;116:1251–1258. doi: 10.1016/j.acthis.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Samarghandian S, Nezhad MA, Mohammadi G. Role of caspases, Bax and Bcl-2 in chrysin-induced apoptosis in the A549 human lung adenocarcinoma epithelial cells. Anticancer Agents Med Chem. 2014;14:901–909. doi: 10.2174/1871520614666140209144042. [DOI] [PubMed] [Google Scholar]
- 42.Brentnall M, Rodriguez-Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biol. 2013;14:32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 44.Mullen P. PARP cleavage as a means of assessing apoptosis. Methods Mol Med. 2004;88:171–181. doi: 10.1385/1-59259-406-9:171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.