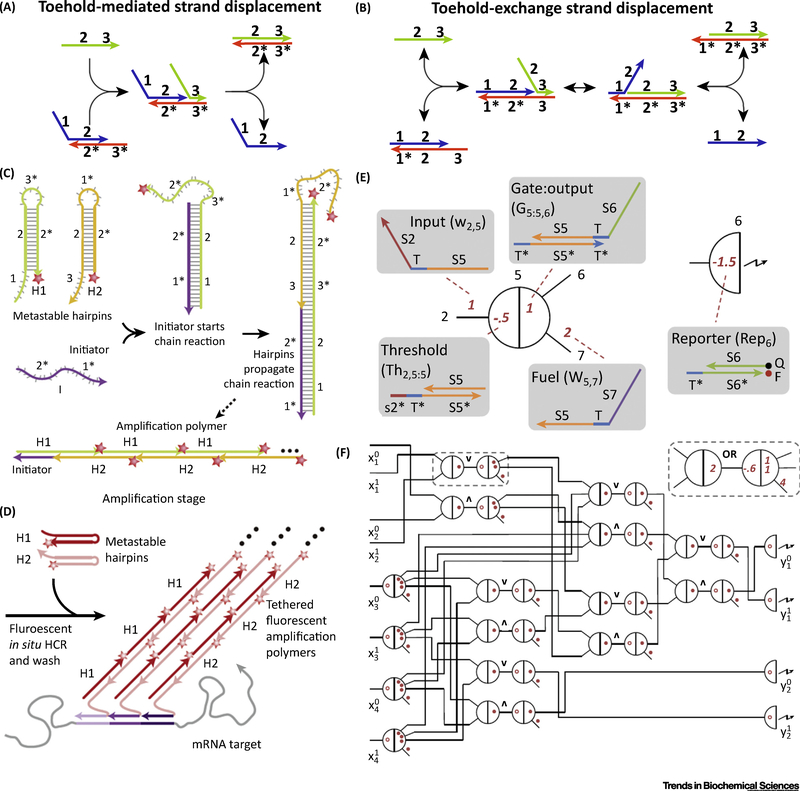
Figure 1. Basic Designs of DNA Dynamic Reaction Networks.
(A) Scheme of toehold-mediated strand displacement reaction. Double-stranded complex (blue and orange) has an overhang domain (3*), known as a toehold. The competitor strand (green) also has a domain 3 which is complementary to 3*. The hybridization of complementary toehold domains acts as the initiation of the following strand migration process, in domain 2/2*. As a result, the competitor strand completely hybridizes with the orange strand while blue strand is released. This process is driven forward by the decrease in free energy on formation of new base pairs. (B) Scheme of toehold exchange strand displacement reaction. The competitor strand (green) hybridizes with the double-stranded complex (blue and orange) via domain 3/3* and a strand migration occurs in domain 2/2*. After the blue strand is released, a new toehold is formed as domain 1, so that the entire process is reversible. (C) Scheme of HCR reaction. In the presence of a single-stranded catalyst, two hairpin structures, H1 and H2, can be opened one by one, forming a long and periodic double stranded DNA chain. (D) Use of HCR as a multiplex amplification strategy for mRNA mapping. (E) See-saw DNA node. (F) Abstract diagram of the seesaw circuit that is equivalent to the square-root digital logic function.