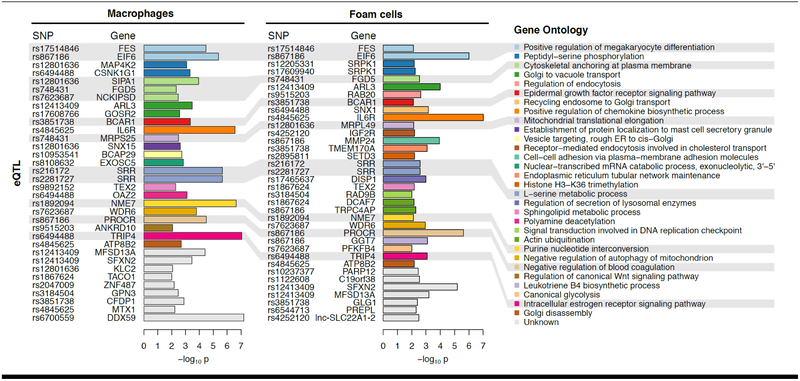
Figure 4. Macrophage-specific association between CAD risk loci and gene expression.
We leveraged the STARNET datasets to perform a pilot analysis on the associations between CAD risk loci (single nucleotide polymorphisms [SNPs]) and gene expression in macrophages and foam cells. Thus, we investigated the 98 CAD-associated GWAS SNPs (129,130), asking whether these SNPs predict macrophage or foam cell gene expression within 1Mb; so-called cis-eQTL. We required that the eQTL had the strongest effect in gene expression from macrophages (n=95) or foam cells (n=80) compared to other tissues in STARNET. This approach identified 29 SNPs that are associated with the expression levels of 52 genes (p<0.01). Thus, 29/98 (~30%) of CAD risk loci are most strongly associated with gene expression in macrophages and/or foam cells. In particular, association with IL6R, NME7, TRIP4, PROCR, and EIF6 was the most significant and detected in both macrophages and foam cells. Candidate genes are color-coded and organized based on Gene Ontology terms according to biological processes of potential relevance to macrophage biology. Further details are provided in the Online Appendix.