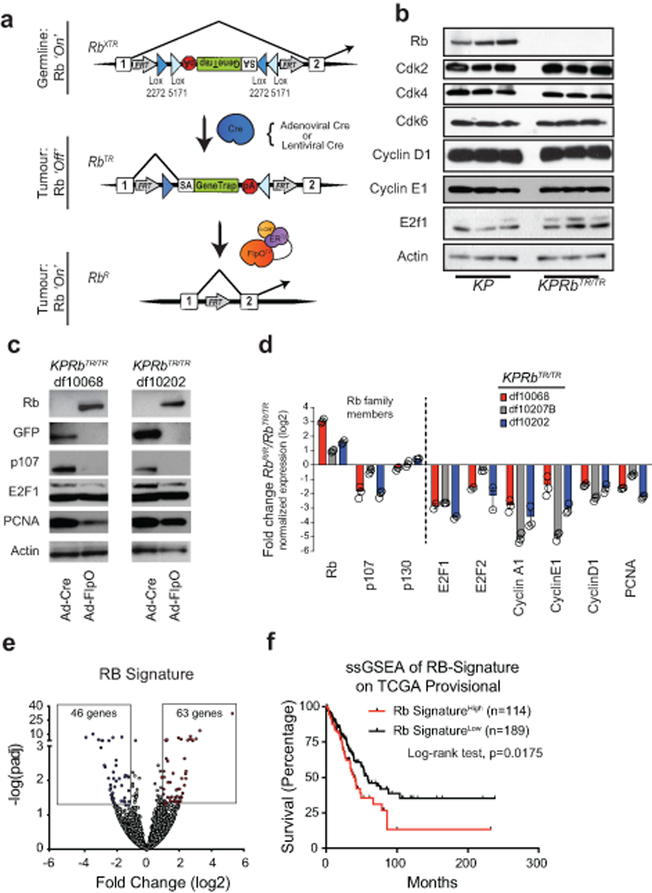
Extended Data Fig. 2: RbXTR allows Cre-dependent inactivation and FlpO-dependent reactivation of RB.
(a) Top: RbXTR (eXpressed): XTR gene trap cassette consists of a splice acceptor (SA), GFP complementary DNA “GeneTrap”, and the polyadenylation transcriptional terminator sequence (pA). Stable inversion is achieved by the use of two pairs of mutually-incompatible mutant LoxP sites (Lox2272 and Lox5171) arranged in the ‘double-floxed’ configuration. In the germline and in normal somatic cells RB expression is normal in RbXTR/XTR mice (Ref. 10). Middle: RbTR (Trapped): Inhalation of Cre-expressing adenoviral or lentiviral vectors induces the permanent conversion of the RbXTR allele to RbTR allele that inactivates RB gene expression. Transcripts are spliced from the upstream exon to the GFP reporter gene and downstream transcription is terminated to functionally inactivate gene function. RB expression is inactivated only in the tumour cells. Bottom: RbR (Restored): The Rosa26FlpO-ERT2 allele enables tamoxifen-dependent conversion of trapped RbTR to its restored RbR allelic state via excision of the gene trap.
(b) Western blot analysis of 3 KP and 3 KPRbTR tumour derived cell lines.
(c) Western blot analysis of 2 KPRbTR tumour derived cell lines treated with Adeno-Cre as a control, or Adeno-FlpO to restore RB expression.
(d) Quantitative RT-PCR analysis of 3 KPRbTR tumour-derived cell lines treated with Adeno-FlpO to restore RB expression. data are normalized to Adeno-Cre treated cells for control. Log2 fold change +/− standard deviation is shown. n=3 technical replicates for each cell line.
(e) Volcano plot of differentially expressed genes from RNA-seq data obtained from KP (n=4) versus KPRbTR/TR (n=4) cell lines. The RB Signature, defined by genes whose expression change is ≥ 2-fold and p-value adjusted for multiple testing ≤ 0.05, is boxed. Statistical significance determined by two-sided Wald’s test using Benjamini-Hochberg correction via DESEQ2.
(f) Kaplan-Meier survival analysis of lung adenocarcinoma patients whose tumours exhibit a high (n=114 patients) or low (n=189 patients) RB Signature. Significance was determined by two-sided Log-rank test (p=0.0175).