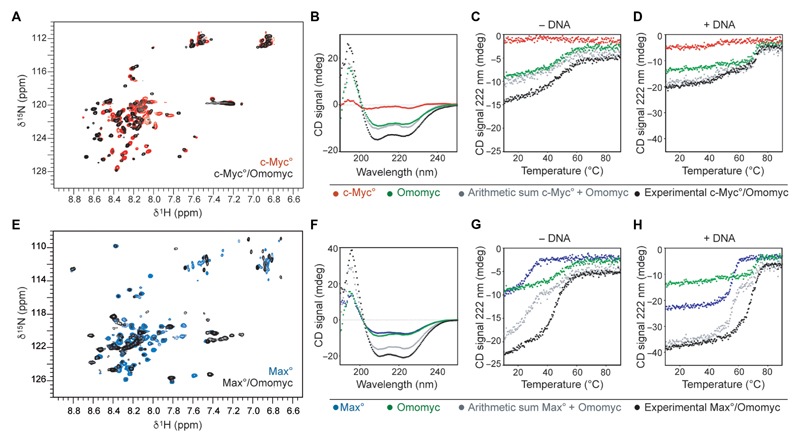
Fig. 1. The Omomyc mini-protein forms highly stable DNA binding homodimers and heterodimers with MAX in solution.
(A) Overlay of the 1H-15N-HSQC of 15N-Myc° in the absence (red) and in the presence (black) of Omomyc. (B) Far–ultraviolet (UV) CD spectra of c-Myc° (red, 8 μM monomer units), Omomyc (green, 8 μM monomer units), the arithmetic sum of both red and green spectra (gray), and the spectrum of an equimolar mix of c-Myc° and Omomyc at a total concentration of 16 μM in monomer units (black) recorded at 25°C. (C) Thermal denaturation of the solutions described in (B). (D) Thermal denaturation of the solutions described in (B) to which equimolar amounts (in dimer units) of an E-box DNA duplex were added. Contribution of the DNA to the denaturation curves was removed. (E) Overlay of the 1H-15N-HSQC of 15N-Max° in the absence (blue) and in the presence (black) of Omomyc. (F) Far-UV CD spectra of Max° (blue, 8 μM monomer units), Omomyc (green, 8 μM monomer units), arithmetic sum of both blue and green spectra (gray), and experimental equimolar mix of Max° and Omomyc at a total concentration of 16 μM in monomer units (black) recorded at 25°C. (G) Thermal denaturation of the solutions described in (F). (H) Thermal denaturation of the solutions described in (F) to which equimolar amounts (in dimer units) of E-box DNA duplex were added. All thermal denaturations were monitored at 222 nm.