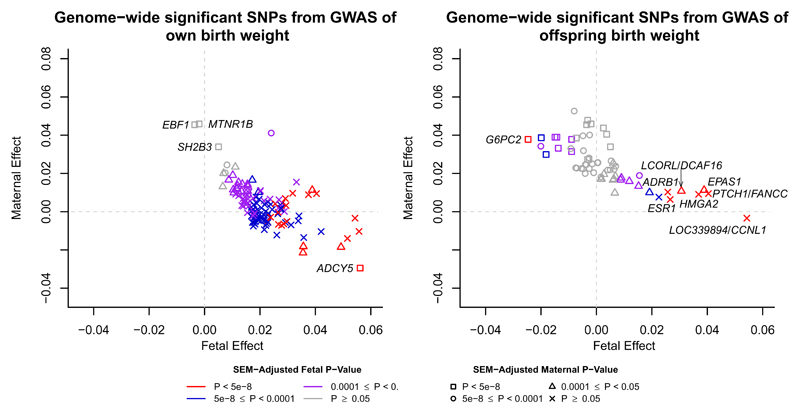
Figure 1. Structural equation modelling (SEM)-adjusted fetal and maternal effects for the 193 lead SNPs that were identified in the GWAS of either own birth weight (left panel) or offspring birth weight (right panel) with minor allele frequency greater than 5%.
The SEM included 85,518 individuals from the UK Biobank with both their own and offspring’s birth weight, 178,980 and 93,842 individuals from the UK Biobank and the EGG consortium with only their own birth weight or offspring’s birth weight respectively. The colour of each point indicates the SEM-adjusted fetal effect on own birth weight association P-value and the shape of each point indicates the SEM-adjusted maternal effect on offspring birth weight association P-value. P-values for the fetal and maternal effect were calculated using a two-sided Wald test. SNPs which are labelled with the name of the closest gene are those which were identified in the GWAS of own birth weight but whose effects are mediated through the maternal genome (left panel) and SNPs that were identified in the GWAS of offspring birth weight but whose effects are mediated through the fetal genome (right panel). SNPs are aligned to the birth weight increasing allele from the GWAS.