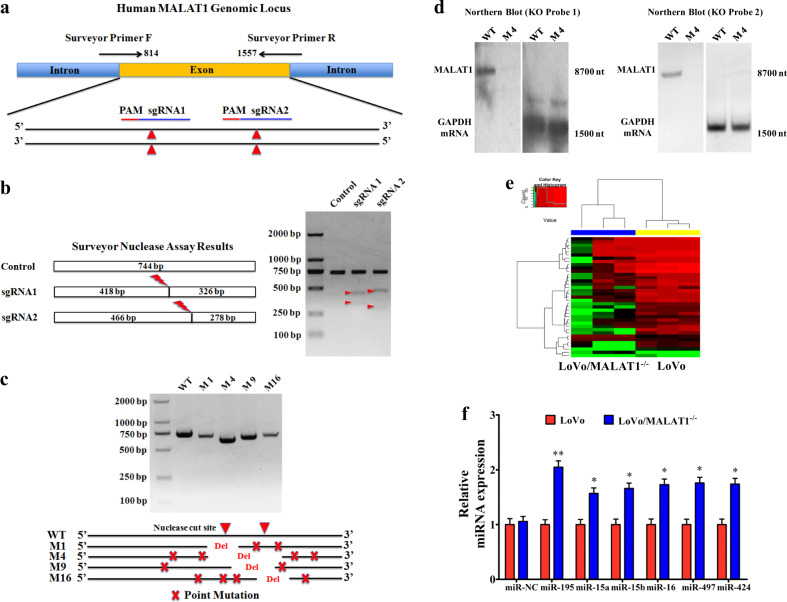
Fig. 2. CRISPR/Cas9-mediated MALAT1 knockout elevates the levels of miR-15s.
a Two CRISPR nuclease sgRNA designs for lncRNA-MALAT1. b Functional validation in HEK293T cells by SURVEYOR Nuclease S Assay. c Validation of CRISPR knockout in single-cell clones by PCR and DNA sequencing. d Northern blot analysis of MALAT1 expression. Twenty micrograms of total RNA from MALAT1 KO LoVo cells (M4) and wild-type LoVo cells were run in agarose/formaldehyde gels. After transferring to nylon membranes, MALAT1 and GAPDH mRNA were detected using antisense oligomer probes (MALAT1 KO Probe 1 targeting the deleted core region, MALAT1 KO Probe 2 targeting the deleted non-core region, and control GAPDH probe) that were end-labeled with 32P. e Cluster analysis of differentially expressed miRNAs in LoVo cells and LoVo/MALAT1−/− cells following by RNA sequencing. Red color represents high expression and green color represents low expression. The color brightness of each unit is associated with differences in multiples (log 2(AR/N). Not all the miRNAs in the figure were labeled. f Validation of the miR-15s levels in LoVo/MALAT1–/–cells by real-time PCR assay. *P < 0.05; **P < 0.01 (t test)