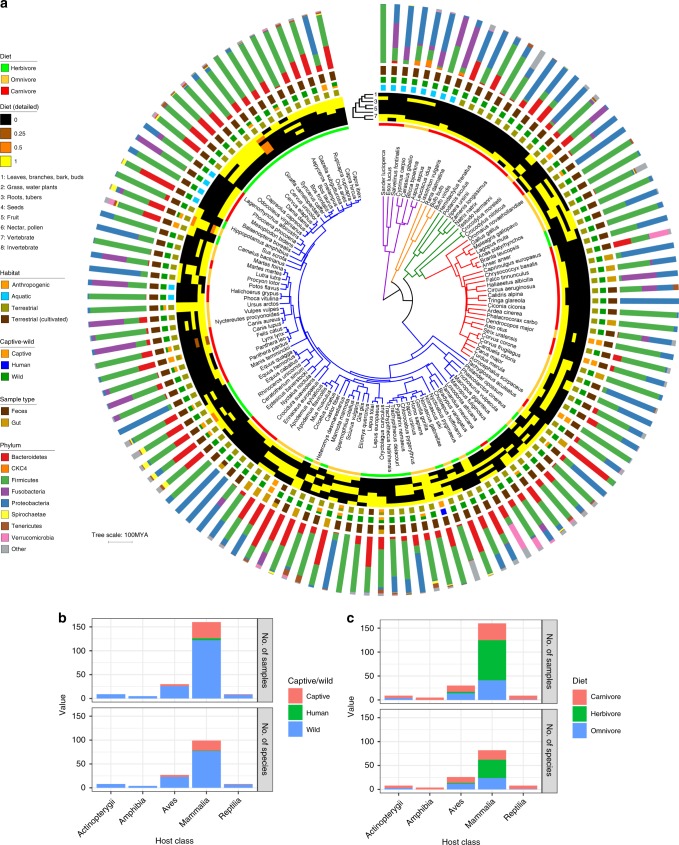
Fig. 1.
Phylum-level grouping of microbiome diversity by host phylogeny and host metadata. a The dated host phylogeny was obtained from http://timetree.org, with branches colored by host class (purple = Actinopterygii; orange = Amphibia; green = Reptilia; red = Aves; blue = Mammalia). From inner to outer, the data mapped onto the tree is host diet (general), host diet (detailed breakdown), host habitat, host captive/wild status, the microbiome sample type, and the relative abundances of microbial phyla in each host. Relative abundances are an estimated average generated via subsampling operational taxonomic units from all samples for each host species (subsampling to 5000 for each host species). Note that “Diet (detailed)” information varies among some individuals, and the values shown are averages of the binary yes/no values (no = 0; yes = 1) for each individual. For example, the Giraffa camelopardalis samples are from two captive and two wild individuals, so the dietary information somewhat differs, resulting in intermediate values (orange). b, c show the number of samples or host species per class colored by captive/wild status or diet, respectively. Source data are provided as a Source Data file