Abstract
The metabolism of the parasite Trypanosoma brucei has been the focus of numerous studies since the 1940s. Recently it was shown, using metabolomics coupled with heavy-atom isotope labelled glucose, that the metabolism of the bloodstream form parasite is more complex than previously thought. The present study also raised a number of questions regarding the origin of several metabolites, for example succinate, only a proportion of which derives from glucose. In order to answer some of these questions and explore the metabolism of bloodstream form T. brucei in more depth we followed the fate of five heavy labelled amino acids – glutamine, proline, methionine, cysteine and arginine – using an LC–MS based metabolomics approach. We found that some of these amino acids have roles beyond those previously thought and we have tentatively identified some unexpected metabolites which need to be confirmed and their function determined.
Keywords: amino acid metabolism, mass spectrometry, parasitic protozoa, trypanosomes
Introduction
Trypanosoma brucei is the causative agent of human and animal African trypanosomiasis. It is a protozoan parasite, transmitted to its mammalian host via a tsetse fly vector. In both hosts the parasite is extracellular, proliferating in the mammalian bloodstream and central nervous system and the tsetse fly midgut from where it migrates through different body tissues before reaching the salivary gland for transmission. T. brucei metabolism is adapted to these disparate environments and significant differences distinguish its metabolism in the different forms of the parasite [1,2].
Within the fly, the main carbon and energy source of the procyclic form trypanosomes is proline, while the bloodstream form parasite uses glucose fed glycolysis as its main energy source [3]. Because of the very high rate of glycolysis, it had long been accepted that the glucose consumed by the parasite was essentially excreted as pyruvate and glycerol. However, the availability of high throughput metabolomics technology, in conjunction with use of isotope labelled substrates, has permitted extensive tracing of glucose metabolism in both the procyclic form [3] and bloodstream form [5] organisms. In bloodstream forms it is clear that glucose enters a plethora of pathways beyond simple glycolysis [5], as is also true for the procyclic form. Unexpected metabolites such as octulose 8-phosphate were identified and labelling patterns revealed a significant role for a succinate shunt generating partially oxidised end products of metabolism. Interestingly, only a proportion of some of these end products, including succinate, is derived from glucose, implying that other sources are used by the cell in their production. It has also been shown in in vitro culture that bloodstream form organisms consume substantial quantities of threonine [6,7] and that a supply of either the aromatic amino acids (tryptophan, tyrosine and phenylalanine) or glutamine was necessary for growth [6]. These appear to be critical cellular sources of nitrogen, fixed via aminotransferase actions. Intriguingly, indole pyruvate, the product of tryptophan has possible immunomodulatory roles [8].
Amino acids clearly play key roles in many aspects of trypanosome metabolism. For example, the unusual metabolite trypanothione (N1,N8-bis(glutathionyl)-spermidine) derives from glutathione (itself consists of the glutamate, cysteine, glycine tripeptide) and the polyamine spermidine, which derives from ornithine via putrescine through sequential addition of aminopropyl groups from decarboxylated S-adenosylmethionine, itself a product of methionine and adenosine. In order to learn more about the utilisation of different amino acids across the trypanosome’s metabolic network we have investigated the distribution of atoms derived from glutamine, proline, methionine, cysteine and arginine using heavy labelled precursors and LC–MS, as these five amino acids will address specific gaps in our current metabolic map as detailed below.
Materials and Methods
Parasite culture and Δargk generation
T. brucei brucei bloodstream form (s427) was cultured in vitro in Creek’s ‘Minimal’ Media (CMM) [6] with 10% FBS Gold at 37°C, 5% CO2. For continuous growth, cell densities were kept between 5 × 104 and 2 × 106 cells/ml and the cell density was checked every 48 h using an improved Neubauer haemocytometer. For growth curves, the cells were counted every 24 h and cell counts performed in triplicate.
The three arginine kinase genes, Tb427tmp.160.4560, Tb427tmp.160.4570, Tb427tmp.160.4590 (Tb927.9.6290, Tb927.9.6250 and Tb927.9.6210 in the 927 reference genome) were knocked out in the bloodstream form s427 strain to obtain strain Δak. The three isoforms are located in an array in the genome (Supplementary Figure S1A), interspaced by a putative gene with unknown function Tb427tmp.160.4580 (Tb927.9.6270). A strategy was designed where the region covering all four genes was replaced with antibiotic resistance cassettes (Supplementary Figure S1B). The 5′ and 3′ flanking regions were amplified from genomic DNA (all primers are detailed in Supplementary Table S1) and cloned flanking a hygromycin or puromycin resistance cassette in vector pTBT [9]. Parasites were transformed in two rounds with NotI-linearised constructs as described previously [10], to replace both allelic regions with the hygromycin and puromycin resistance cassettes through homologous recombination. Positive clones were selected with 2.5 μg/ml hygromycin and 0.2 μg/ml puromycin, and correct integration of the resistance cassettes was confirmed by PCR (Supplementary Figure S1C). Absence of additional isoforms of TbAK in the Δak strain was validated by PCR, following standard procedures [11] and probed with Easytides 32P-ATP (Perkin Elmer) incorporated into TbAK using the Stratagene Prime-it kit.
Intracellular metabolite extraction from parasites
Trypanosomes were grown to mid-log phase and a sample volume equivalent to 5 × 107 cells was rapidly cooled to 4°C by submerging a 50 ml Falcon tube containing the parasites in a dry ice/ethanol bath. Samples were kept on ice (or 4°C) from this step onwards. Samples were centrifuged at 1250g for 10 min, and most supernatant, except 1 ml, was removed. The pellet was resuspended in the remaining 1 ml of medium. This was transferred to an Eppendorf tube and briefly centrifuged at 1250g to completely remove the supernatant. The cell pellet was washed in cold 1× PBS and metabolites extracted by resuspending in 100 μl chloroform:methanol:water (ratio1:3:1 v/v/v) with internal standards and by vigorously shaking for 1 h at 4°C. Samples were centrifuged at 16,000g for 10 min and the supernatant collected and stored at −80°C under argon.
13C-labelled tracking
T. b. brucei strain 427 was grown in CMM + 10% FBS Gold (PAA, Piscataway, NJ) at 37°C, 5% CO2 for U-13C-labelled tracking. For l-methionine studies, a 1:1 mix of unlabelled and U-13C l-methionine (50 μM l-methionine and 50 μM U-13C l-methionine) was added to the parasite culture medium (starting density of 2 × 104 cells/ml). For l-glutamine and l-cysteine studies, a 1:1 mix of unlabelled and U-13C glutamine or cysteine (500 μM unlabelled and 500 μM U-13C) was added to the parasite culture medium. For l-proline and l-arginine, 100% labelled compound was used at a concentration of 200 μM as significant levels of unlabelled l-proline and l-arginine are present in the serum. Cells were incubated at 37°C, 5% CO2 for 48 h, and metabolites extracted as described above. Samples were prepared in triplicate (biological replicates) and for every labelled setup a control with the equal concentration of unlabelled compound was set up, also in triplicate. Fresh medium and spent medium controls (for the l-methionine, l-proline and l-arginine experiments) were also collected and 5 μl were added to 100 μl extraction solvent (CMW 1:3:1) prior to analysis.
Labelled compounds were obtained from Cambridge Isotope Laboratory, Inc.: (l-methionine, 13C5, enrichment 99%, cat: CLM-893-H-0.1; l-proline, 13C5, enrichment 99%, cat: CLM-2260-H; l-arginine, 13C6, enrichment 99%, cat: CLM-2265-H-0.1; l-cysteine, 13C3, enrichment 99%, cat: CLM-4320-H-PK; l-glutamine, 13C5, enrichment 99%, cat: CLM-1822-H-PK).
Sample preparation for pH stress
T. b. brucei strain 427 wild type (WT) and arginine kinase knockout (Δak) cells were cultured in CMM with 10% FBS Gold at 37°C, 5% CO2 as described above and allow them to reach a density of 2 × 106 cells/ml. After a density of 2 × 106 cells/ml was achieved, WT and Δak cells were centrifuged at 1250g for 10 min and the cells were then resuspended in CMM at pH 8.7 (pH stress) and pH 7.4 (controls) and incubated for 120 min.
The cells were then extracted as described in the section ‘Intracellular metabolite extraction from parasites’. Equal volumes from all the samples were mixed to create a quality control (QC) sample in order to assess instrument performance. All samples were prepared in four biological replicates.
Liquid chromatography–mass spectrometry
The sample platform chosen for this project was liquid chromatography coupled with mass spectrometry. All samples were separated on a Dionex Ultimate 3000 RSLC liquid chromatography system (Dionex, Camberley, Surrey) with ZIC-pHILIC 150 × 4.6 mm, 5 µm column (Merck Sequant) prior to mass detection on an Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Hemel Hempstead, U.K.). LC–MS analysis was performed in positive and negative electrospray mode (m/z 70–1400, resolution 50,000), using 10 μl injection volume and a flow rate of 300 μl/min. For HPLC gradient, ZIC-pHILIC solvent A was 20 mM ammonium carbonate in H2O, and solvent B was 100% acetonitrile. A linear gradient of the mobile phases was applied, in which samples were eluted from 80% B to 5% B over 15 min, followed by another linear gradient from 5% B to 80% B over 2 min, and 80% B was held for 7 min for re-equilibration. With each experiment a set of authentic standards was run prior to the sample set.
Data processing and analysis
Raw LC–MS data files were converted to mzXML using ProteoWizard msconvert [12]. Data were processed using XCMS [13] for peak picking and mzMatch for peak grouping, filtering and putative identifications [14]. Metabolite identification was performed by matching accurate masses and retention times of authentic standards. Level 1 metabolite identification according to the metabolomics standards initiative [15,16], but when standards were not available masses and labelling patterns were used, hence these identifications should be considered as putative (Level 2 identification). Isotopic profiles were analysed using mzMatch-ISO [4].
In order to visualise metabolic differences between control and high pH treated cells, orthogonal partial least squares-discriminant analysis (OPLS-DA) was carried out by SIMCA-P 14.1 (Umetrics AB, Umea, Sweden). Mass ions (m/z) that were changed significantly between the two groups were selected by variable importance in projection values (VIP) where the values greater than one were determined as potential key markers. Additionally, univariate t-test analysis was performed using a web-based analysis tool, Metaboanalyst [17] to cross check significant difference between control and treated cells.
The mzXML files of all the experiments described in the present study are available in the Metabolights database at https://www.ebi.ac.uk/metabolights/MTBLS624.
Results and discussion
A recent study by Creek et al. [5] showed that glucose was a source of carbon for many pathways beyond glycolysis. The present study also raised new questions, as many of these pathways only use glucose for part of their carbon needs. To extend the measured metabolic map of bloodstream form T. brucei, we followed the fate of five key amino acids, arginine, cysteine, glutamine, methionine and proline inside the cells, using U-13C-labelled amino acids and LC–MS.
Glutamine
As previously reported [6,7,18], glutamine is consumed by bloodstream form T. brucei from CMM in large quantities where it has been proposed it mainly serves as a key amino group donor. 2-Oxoglutarate and 2-oxoglutaramate as well as some glutamate are secreted into the medium as bloodstream forms grow [6,18]. Furthermore, significant quantities of succinate are secreted by bloodstream forms [6,7,18], of which only 52% comes from glucose via the essential phosphoenolpyruvate carboxykinase [5]. The remaining succinate must be produced from another carbon source, and glutamine-derived 2-oxoglutarate is a likely candidate. To test this hypothesis, we incubated T. brucei bloodstream form for 48 h in CMM with 50% U-13C glutamine and then analysed the intracellular metabolome using LC–MS (see Materials and Methods).
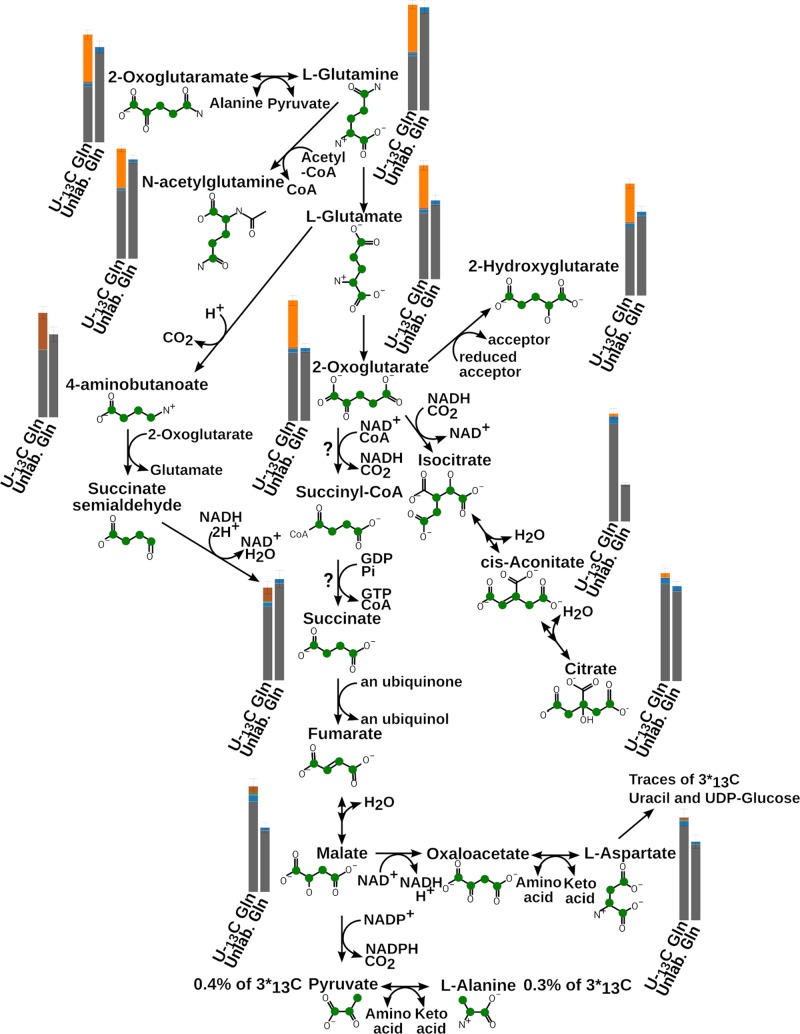
The results (see Figure 1) reveal that 45% of intracellular glutamine is 5 carbon labelled (additional unlabelled glutamine from the serum likely having a small dilution effect). Acetylated glutamine is also present (N-acetylglutamine labelled at 37 ± 8%). Glutamate is 37.5 ± 0.3% fully labelled and 2-oxoglutarate 39.0 ± 0.3%; confirming that the primary source of glutamate and 2-oxoglutarate inside the cell is medium-derived glutamine. Labelling can also be seen in glutathione (39.4 ± 0.1%) and trypanothione disulphide (39 ± 1%), as expected (see Figure 2).
Figure 1. Glutamine labelled experiments results.
The bar charts show the measured proportions of the different number of 13C: grey represents fully unlabelled metabolite, blue 1 carbon 13C, brown 4 carbons 13C and orange 5 carbons 13C. The green dots on the molecules represent the theoretical individual 13C starting with a fully labelled precursor following this pathway.
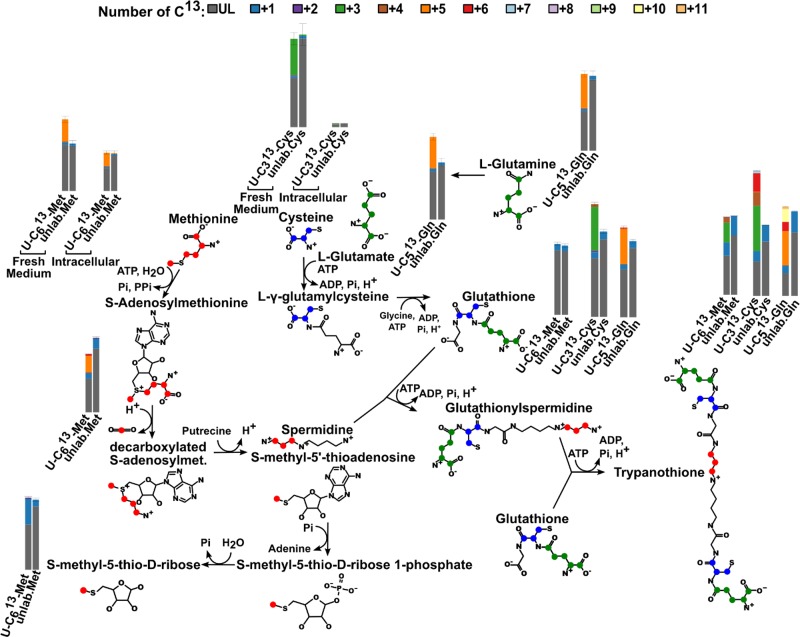
Figure 2. Trypanothione biosynthesis pathway as seen using 50% U-13C glutamine, cysteine or methionine.
The bar charts show the measured proportions of the different number of 13C (colour code is shown at the top of the figure). The green dots on the molecules represent the theoretical individual 13C coming from fully labelled glutamine, blue carbons come from fully labelled cysteine and red from fully labelled methionine.
Although a canonical glutaminase (EC 3.5.1.2) encoding gene cannot be found in the T. brucei genome [19], several reactions could be responsible for producing glutamate from glutamine, for example: (i) a GMP synthase (EC. 6.3.5.2, gene Tb927.7.2100) that converts XMP to GMP uses glutamine as an amino group donor [20], (ii) a glutamine-dependent carbamoyl-phosphate synthase (EC 6.3.5.5, putative gene Tb927.5.3800, syntenic orthologue of the identified Leishmania mexicana and Trypanosoma cruzi genes [21] also uses glutamine as an amino group donor to produce carbamoyl-phosphate from hydrogen carbonate [22] and is also essential [23], (iii) an asparagine synthase (Tb927.7.1110) can use either ammonia (EC 6.3.1.1) or glutamine to produce asparagine from aspartate [24].
Many reactions can produce 2-oxoglutarate from glutamate. For example, in T. brucei, glutamate dehydrogenase [25]. Two isoforms of aspartate aminotransferase (EC 2.6.1.1, mitochondrial isoform gene Tb927.11.5090, cytosolic isoform gene Tb927.10.3660) are also present in bloodstream forms [27] and alanine aminotransferase (EC 2.6.1.2, gene Tb927.1.3950) is expressed at high levels [28]. The putatively identified 2-oxoglutaramate is present in the labelled form at high levels (43.5 ± 0.1%) and is probably derived from the glutamine aminotransferase (EC 2.6.1.15, gene Tb927.10.11970) that produces 2-oxoglutaramate and alanine from glutamine and pyruvate [28].
The total succinate pool is present with 14.9 ± 0.1% containing 4 labelled carbons indicating that part of the total succinate pool is derived from glutamine, most likely via 2-oxoglutarate. Indeed, enzyme activities of 2-oxoglutarate dehydrogenase (EC 1.2.4.2) and acetyl:succinate CoA-transferase (EC 2.8.3.18, gene Tb927.11.2690) have been measured in bloodstream forms, albeit at lower levels than in procyclics [29,30]. In addition to succinate, 4 carbon labelled malate is also present (6.2 ± 0.1% of the total measured malate) presumably due to a reverse flux from succinate [7]. Malate then contributes to the low levels of labelled aspartate (2.6 ± 0.2% of the total) via malate dehydrogenase (EC 1.1.1.299, cytosolic isoform gene Tb927.11.11250) [29,31,32] and aspartate aminotransferase. The promiscuous action of malate dehydrogenase on 2-oxoglutarate [33] also produces 5 carbon labelled 2-hydroxyglutarate [34] (34.1 ± 0.5% of the total) which is transformed to 2-oxoglutarate by 2-hydroxyglutarate dehydrogenase (EC 1.1.99.2, putatively annotated gene Tb927.10.9360).
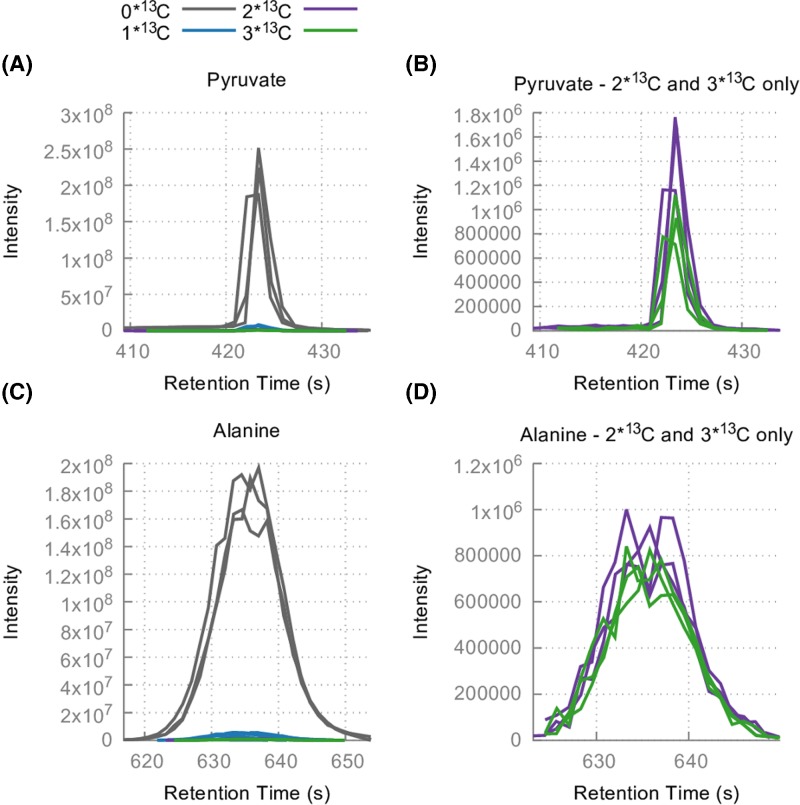
Bloodstream form T. brucei contain large concentrations of pyruvate and alanine in their cytoplasm [18,35], most of which is derived from glucose (close to 100% in [5], although labelling experiments show that a small but significant quantity of the total (0.4 ± 0.02% of the pyruvate and 0.3±0.03% of the alanine) is labelled from glutamine (see Figure 4). Malic enzyme (EC 1.1.1.40, mitochondrial isoform Tb927.11.5450, cytosolic isoform Tb927.11.5440), the activity of which has been measured in bloodstream forms [29,33,36,37] could produce this pyruvate and subsequently alanine via alanine aminotransferase and glutamine aminotransferase.
Figure 4. Pyruvate (A and B) and alanine (C and D) as seen in the 50% U-13C glutamine experiment.
Small amounts of 5 carbon labelled citrate/isocitrate (these two isomers cannot be distinguished on this platform) and cis-aconitate (4.0 ± 0.6% of citrate and less than 1% of cis-aconitate) also derive from the labelled glutamate, as also seen from labelled glucose (approx. 11% of citrate/isocitrate is found labelled from glucose and 5% of cis-aconitate) [5]. Isocitrate dehydrogenase can produce isocitrate from 2-oxoglutarate (EC 1.1.1.42, gene Tb927.11.900) and the protein is identified in bloodstream forms [33,37,38] despite its activity being below the detection threshold when assayed [30]. Aconitase activity was detectable [30] and the protein detected [39] (EC 4.2.1.3, gene Tb927.10.14000), thus explaining the labelled cis-aconitate and citrate. The proportion of the labelled derivatives is low when compared with unlabelled due to the parasites accumulating large quantities of citrate from medium [38]. It is clear that glutamine can be used to fuel various pathways in bloodstream form T. brucei, with a primary role in provision of amino groups to the nitrogen pool of the parasites.
Proline
Proline is the primary source of carbon and energy in procyclic form trypanosomes and critical to viability in the tsetse fly [40]. Enzymes involved in its catabolism are substantially less abundant in the bloodstream form and it has been assumed that proline has little metabolic use to these cells [41]. Given that the presence of peptidases in the medium increases the concentration of unlabelled proline over time [18], the cells were incubated in medium containing 200 µM proline 100% U-13C-labelled. After 48 h 49 ± 1% of the proline inside cells was labelled (unlabelled proline coming from the serum causes the dilution) while very small quantities (less than 1%) of succinate, malate, 2-oxoglutarate and glutamate were labelled (4 carbons for succinate and malate, 5 for glutamate and 2-oxoglutarate) (see Supplementary File S4) confirming that these cells barely utilise l-proline through catabolic pathways although they maintain low level activity of the enzymes necessary to do so.
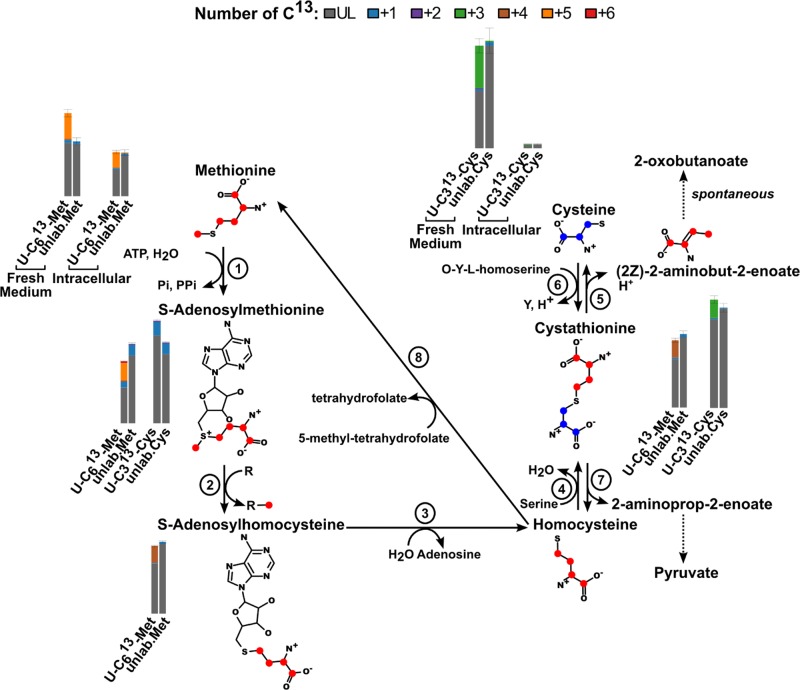
Methionine
Methionine has several roles of fundamental importance in trypanosomes. For example, it is the primary source of methyl groups in a multitude of reactions when converted to S-adenosylmethionine (AdoMet) and also of aminopropyl groups in growing polyamine chains after production of the decarboxylated form. Creek et al. previously revealed that S-adenosyl-l-methionine is 5 carbon labelled when using U-13C glucose due to labelling of the ribose moiety of the adenosyl group (76% labelled in [5]. Decarboxylated Adomet is used exclusively for the conversion of putrescine to spermidine which creates S-methyl-5′-thioadenosine (MTA) once the aminopropyl group has been transferred to the growing polyamine chain. In many cell types, MTA is recycled to methionine and this was presumed to occur also for bloodstream form T. brucei [42]. However, Creek et al. showed this is not the case. Labelling with 50% U-13C l-methionine and identifying its metabolic products both within the parasites and their medium has given more insight into the fate of MTA, whose accumulation is generally toxic [43,44].
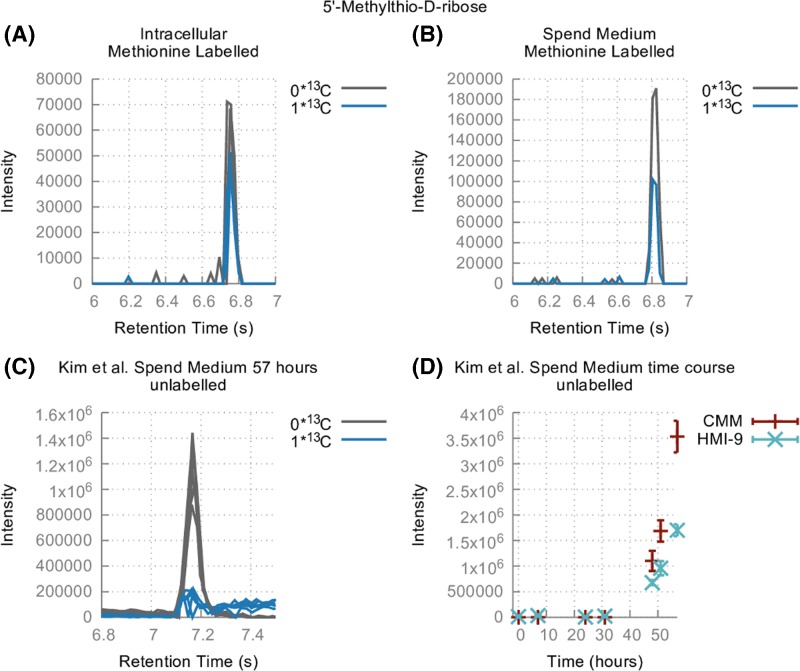
MTA is not converted to l-methionine (see Figure 2) using the classical methionine cycle since this pathway would create 1 carbon labelled l-methionine from U-13C l-methionine and no excess 1 carbon labelled product is detected when compared with the unlabelled control. 35 ± 6% of intracellular methionine is labelled with 5 carbons for 48 h and 28.2 ± 0.1% of S-adenosyl-l-methionine is also 5 carbon labelled. A peak corresponding to the mass of MTA was labelled with a single carbon (36 ± 2% 1 carbon 13C-labelled). This peak is only detected within cells and not in the spent medium. By contrast, a low intensity peak with a significant proportion of 1 carbon label corresponding to the mass of methylthioribose was identified in spent medium (see Figure 3A,B). Since this peak was of low intensity we investigated its appearance in spent medium in a time course experiment [18]. This metabolite accumulates in the medium proportionally to the cell density (see Figure 3C,D). This indicates that bloodstream form T. brucei metabolise methionine via AdoMet to MTA which is converted to methylthioribose 1-phosphate (EC 2.4.2.28, gene Tb927.7.7040) [45], then dephosphorylated to methylthioribose which is secreted from the cell. It appears likely, therefore, that T. brucei has evolved a mechanism to detoxify MTA generated during polyamine biosynthesis by converting to MTR which is subsequently secreted from the cell, as is also the case in E. coli [46].
Figure 3. Putative methylthio-d-ribose in the U-13C methionine experiment (A and B) and in Kim et al. [18] spend medium experiment (C and D).
A metabolite with mass consistent with methionine’s ketoacid derivative, 2-oxo-4-methylthiobutanoic acid, was also identified. This metabolite is present with 33 ± 1% 5 carbon labelled both intracellularly and in the spent medium, indicating it arises from the transamination of methionine and not MTA recycling (in which case it would be 1 carbon 13C-labelled). This transamination is probably catalysed by aspartate aminotransferase [26].
During spermidine synthesis, three carbons in the aminopropyl group released from decarboxylated AdoMet are labelled and since spermidine is then incorporated into trypanothione we identify 3 carbon labelled trypanothione (see Figure 2).
S-Adenosyl-l-methionine is also a major methyl group donor. Methylation reactions convert S-adenosyl-l-methionine into S-adenosyl-l-homocysteine, which we detect with 24 ± 4% 4 carbon labelled (see Figure 5). Several metabolites can also be detected as methylated derivatives in this experiment (Table 1 gives a list of the labelled metabolites detected, the number of labelled carbons and their potential functions).
Figure 5. S-Adenosylhomocysteine recycling pathway.
The bar charts show the measured proportions of the different number of 13C (colour code is shown at the top of the figure). The blue dots on the molecules represent the theoretical individual 13C coming from cysteine and red from methionine.
Table 1. Methylated metabolites detected in the U-13C methionine experiment.
| Metabolite | Source of identification | Carbons 13C-labelled | Putative function | Putative gene involved | Reference |
|---|---|---|---|---|---|
| Nγ-Monomethyl-l-arginine | Mass and labelling | 1 | Protein/histone methylation | Tb927.1.4690 (type I) Tb927.5.3960 (type I) Tb927.10.640 (type II) Tb927.7.5490 (type III) Tb927.10.3560 | [48,50] – five putative arginine protein methyltransferases: type I: [51,52], type II: [53], type III: [54] (histone: [55,56]) |
| Nω, Nω′-Dimethyl-l-arginine | Mass and labelling | 1 and 2 | Protein/histone methylation | Tb927.1.4690 (type I) Tb927.5.3960 (type I) Tb927.10.640 (type II) Tb927.10.3560 | See Nγ-monomethyl-l-arginine |
| N6-Methyl-l-lysine | Mass and labelling | 1 | Protein/histone methylation | Tb927.8.1920, Tb927.1.570 | [59–62] |
| N6,N6,N6-Trimethyl-l-lysine | Mass and labelling | 1, 2 and 3 | Protein/histone methylation | Tb927.8.1920, Tb927.1.570 | See N6-methyl-l-lysine |
| Methylguanine | Mass and labelling | 1 | RNA cap | Tb927.11.4890, Tb927.10.7940 | [59,60] |
| Nα-Methylhistidine | Mass, retention time (match standard) and labelling | 1 | Histidine methylation seen in histones and other proteins in Leishmania | [61] (Leishmania) |
The product of these methylation reactions, S-adenosyl-l-homocysteine is another toxic thiol which needs to be removed from the system [47]. The classical pathway of detoxification involves a salvage pathway that ultimately produces l-cysteine (see Figure 5). Labelled carbons from the methionine precursor are first passed on to homocysteine (undetected) via adenosylhomocysteinase (EC 3.3.1.1, putative gene Tb927.11.9590 [48]), then cystathionine via cystathionine β-synthase (EC 4.2.1.22, putative genes Tb11.02.5400b and Tb11.02.5400 – orthologue of the identified T. cruzi gene [49]) and finally 2-aminobut-2-enoate via cystathionine γ-lyase (EC 4.4.1.1, putative genes Tb927.9.12320 and Tb11.v5.0869, orthologues of the identified Leishmania major gene [50] and oxobutanoate). Of these four metabolites, we detect cystathionine which is indeed 26 ± 1% 4 carbon labelled. This supports the presence of this salvage pathway as previously indicated by Bacchi et al. using 35S labelled methionine [51]. However, since bloodstream form T. brucei consume large quantities of cysteine from their medium [6,52] the salvage pathway is probably more important in preventing accumulation of toxic S-adenosyl-l-homocysteine and l-homocysteine than in providing cysteine which needs to be present in the medium for the parasite to be able to grow [52].
Homocysteine can also be reconverted to methionine by 5-methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase (EC 2.1.1.14, putative gene Tb927.8.2610) or homocysteine S-methyltransferase (EC 2.1.1.10, putative gene Tb927.1.1270) [53]. The first reaction would produce four carbons 13C-labelled methionine. However, whilst a metabolite of this mass is detected, it is also found at the same level in fresh medium containing U-13C methionine, indicating that it is a contaminant of the label stock and not a product of cellular metabolism. The second reaction, homocysteine S-methyltransferase [53], would produce U-13C methionine if the co-substrate S-methyl-l-methionine is also derived from S-adenosyl-l-methionine. However, this pathway would also produce additional S-adenosyl-l-homocysteine and therefore would not reduce the concentration of this metabolite.
This data indicate that methionine is a key donor of methyl groups following conversion to AdoMet and aminopropyl groups in polyamine biosynthesis. In each case, potentially toxic by-products are produced, and it appears that the parasite has evolved a mechanism to detoxify MTA, by conversion in two steps to MTR which is secreted, and S-adenosyl-l-homocysteine through conversion to cysteine.
Cysteine
26 ± 1% of cystathionine is labelled when U-13C methionine is added to culture medium. As 35 ± 6% of total intracellular methionine is labelled after 48 h, approx. 74% of cystathionine would appear to come from the S-adenosyl-l-homocysteine recycling pathway. This indicates that one or more other pathways must account for the remaining 26%. The most likely additional source of cystathionine is cysteine, which is consumed in large quantities by bloodstream form T. brucei [6]. Cells were grown for 48 h in CMM containing 50:50 U-13C cysteine: unlabelled cysteine to follow the fate of cysteine inside the cells.
Labelled cysteine is seen within cells, but the peaks are of too low intensity to allow quantification of relative proportions of labelled and unlabelled versions. Around 20% of cystathionine is labelled at three carbons, indicating it is derived either from the reverse reaction of cystathionine γ-lyase (Figure 5, reaction 5) or from cystathionine γ-synthase (Figure 5, reaction 6) [54]. No orthologues of known variants of the latter enzyme are found in the genome although the presence of a non-canonical enzyme cannot be excluded. Cysteine-derived carbons could be transferred to 2-aminoprop-2-enoate by a cystathionine β-lyase (EC 4.4.1.8, protein sequence very similar to cystathionine γ-lyase) [54]. 2-aminoprop-2-enoate would in turn be converted to pyruvate. However, 2-aminoprop-2-enoate cannot be detected and we cannot see any trace of labelled pyruvate derived from cysteine in this dataset making its presence unlikely. Only growing cells in the presence of 34S labelled cysteine would allow us to prove or disprove this hypothesis.
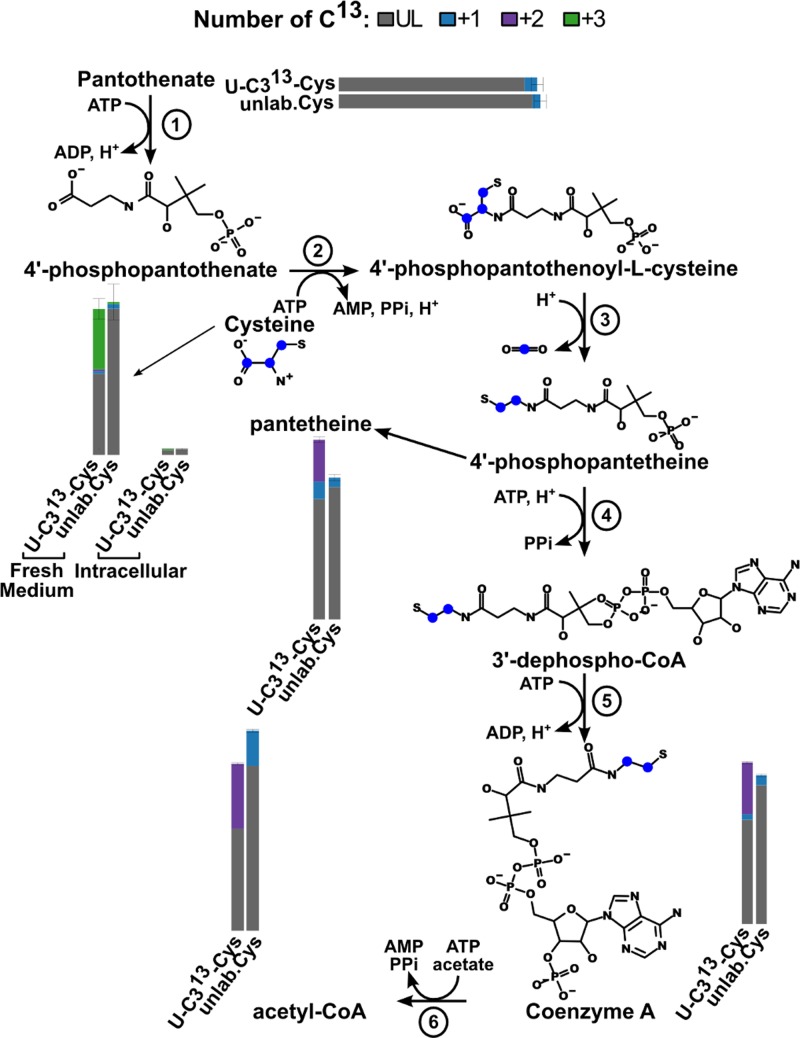
Cysteine is also essential to produce glutathione, trypanothione and coenzyme A (CoA). As expected, glutathione is present with 39 ± 4% 3 carbon 13C-labelled and trypanothione with 38 ± 5% 3 carbon 13C-labelled and 16 ± 2% 6 carbon 13C-labelled (see Figure 2). CoA is most likely produced from pantothenate taken up from the medium (see Figure 6), even though its consumption rate could not be measured due to the small amount likely required by the cells [18]. Indeed, we can detect coenzyme A and acetyl-CoA with 2 carbon labelled as the biosynthesis pathway predicts. We also detect a 2 carbon labelled metabolite the mass of which is consistent with pantetheine, a likely fragment of phosphopantetheine. The first four reactions of CoA synthesis are catalysed by enzymes that are all putatively annotated in the T. brucei genome but have never been tested (see Figure 6). The fifth reaction, dephospho-CoA kinase (EC 2.7.1.24, putative gene Tb927.6.710) has been localised to glycosomes [45]. Coenzyme A can then be converted to acetyl-CoA either via the pyruvate dehydrogenase complex (EC 1.2.1.-) or via the acetyl-CoA synthetase (EC 6.2.1.1, Tb927.8.2520 [55]) as shown by Mazet et al. [7].
Figure 6. Coenzyme A biosynthesis pathway.
The bar charts show the measured proportions of the different number of 13C (colour code is shown at the top of the figure). The blue dots on the molecules represent the theoretical individual 13C coming from cysteine.
Arginine
Arginine plays several important roles in cellular physiology in many systems. It is, for example, a precursor to polyamine synthesis in most systems, producing ornithine via arginase. Furthermore, it generates citrulline and nitric oxide via the nitric oxide synthase (NOS). A gene encoding an enzyme of the arginase family was found in T. brucei [56] but the protein lacked arginase activity [57]. Instead it seems that ornithine is accumulated from outside the cells [58]. Here we investigated the fate of arginine in trypanosomes by growing them in presence of 200 µM U-13C arginine in CMM.
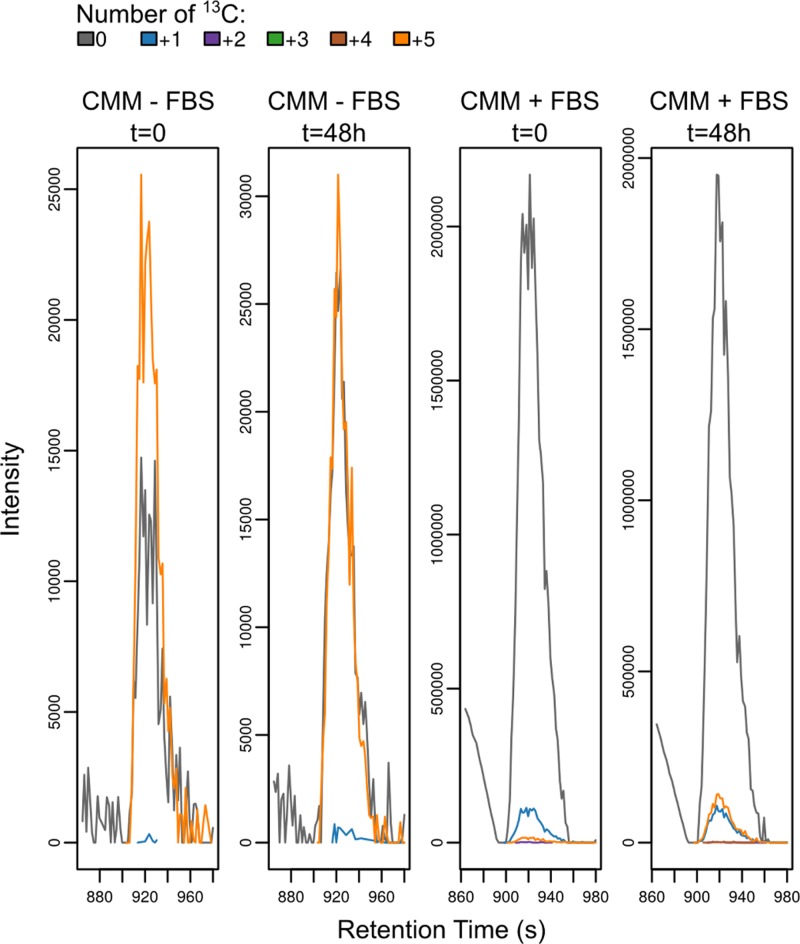
By the time a steady state was reached, 40 ± 1% of intracellular arginine is fully labelled (a dilution effect is most likely caused by the peptidases secreted in the medium [18] freeing additional unlabelled arginine from serum proteins). 11.2 ± 0.5% of intracellular ornithine was labelled with 5 13C carbons, consistent with it being produced by arginase, previously reported as missing from trypanosomes [57]. However, the same proportion of labelled ornithine can be seen in the spent medium (but not in the fresh medium), raising the possibility that some arginine is converted to ornithine in the medium, possibly by arginase in the serum. To test this hypothesis, we added 200 µM of U-13C arginine to the medium with or without FBS and without adding cells. After 48 h we can see an increase in the labelled ornithine produced only with added FBS. This reveals that ornithine is indeed produced from arginine when no cells are present (see Figure 7) hence the labelled ornithine observed inside trypanosomes appears to be created by arginase present in serum and then secondarily accumulated by trypanosomes.
Figure 7. Peak of ornithine as seen in the medium (CMM) when arginine is fully labelled, with or without FBS but no cells.
(Intensity of the peak shown as a function of retention time).
The methionine labelling experiment revealed mono and dimethylarginine present with 22 ± 2% and 26 ± 1% of their total being labelled with 6 carbons. A small proportion of fully labelled citrulline (2.8 ± 0.7%, the fragmentation of this peak was checked and is consistent with citrulline) is also detected in cells but very little in the spent medium. Since T. brucei does not have NOS activity [59] it is likely that the citrulline derives from dimethylarginine via dimethylarginase activity although the canonical enzyme is not found in the genome (dimethylated arginines are found on histones for example).
A metabolite with a mass corresponding to 5-guanidino-2-oxo-pentanoate is also detected (37 ± 1% of its total quantity labelled). This metabolite would be produced by an aminotransferase reaction using arginine as the nitrogen donor.
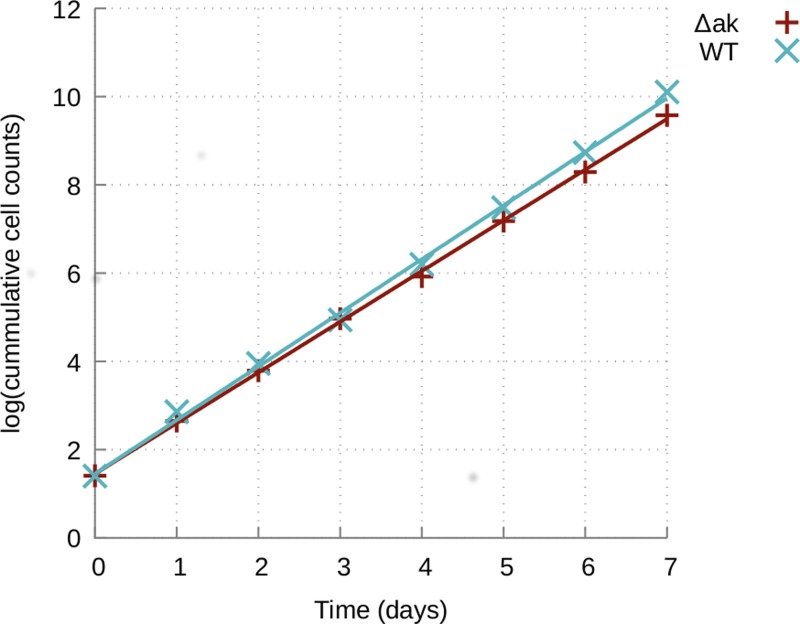
Finally, a metabolite with a mass consistent with l-arginine phosphate is also detected (37 ± 1% being 6 carbon labelled) which is consistent with the presence of arginine kinase (AK) in these cells (EC 2.7.3.3, gene Tb927.9.6290, Tb927.9.6250 and Tb927.9.6210) [60], localised to the glycosome, cytosol and flagellum, respectively [60]. In order to confirm that the arginine phosphate detected was indeed derived from AK, we knocked out all three highly similar isoforms of the gene in the BSF of T. brucei (see Supplementary Figure S1). The Δak strain was viable and had a similar growth rate to the wild type (Figure 8). In addition, untargeted metabolite profiling of the Δak line showed that arginine phosphate was no longer present, confirming that l-arginine is the sole source of arginine phosphate in these cells. Interestingly the level of l-arginine was also reduced by 1.7-fold, but no significant global metabolic changes were observed between Δak and wild type cells.
Figure 8. Growth of wild type and Δak cells.
Although no significant metabolic changes were caused by the absence of AK genes, arginine phosphate is believed to play a critical role as an energy reserve by transferring the high-energy phosphate of arginine-phosphate back to ADP to form ATP when it is required quickly, for example under severe stress conditions such as starvation, nutritional and pH stress in trypanosomes [61,62].
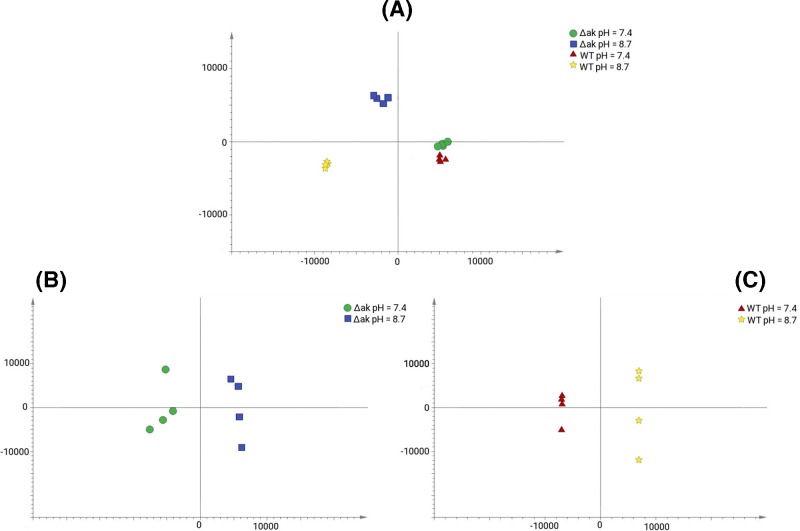
The role of AK in trypanosome survival at high pH was also investigated. The survival rate of Δak and WT cells grown at pH 7.4 (normal growth condition) and 8.7 (high pH condition) was investigated by counting the number of cells surviving after 72 h of cell culture at each pH condition. The Δak cells showed a 52% decrease in survival capability at pH 8.7 compared with the cells grown at pH 7.4 (Δak at pH 7.4, (5.78 ± 0.25) × 106 cells/ml; AK KO at pH 8.7, (3.37 ± 0.12) × 106 cells/ml) whereas the WT cells showed only a 22% decrease in cell density growing at pH 8.7 (WT cells at pH 7.4, (6.27 ± 0.1) × 106 cells/ml; WT cells at pH 8.7, (5.54 ± 0.47) × 106 cells/ml) (Table 2). This indicates that arginine kinase improves the ability of WT cells to resist high pH stress conditions. In order to further investigate the effects of high pH growth conditions at the molecular level, extracts from the Δak and WT cells grown under pH 7.4 and 8.7 were analysed by LC–MS. OPLS-DA was then performed to determine specific metabolites affected by the different growth conditions. As can be seen in Figure 9A, a clear separation was observed between the normal growth and high pH growth conditions (Δak grown at pH 7.4 (A74) vs pH 8.7 (A87); WT grown at pH 7.4 (W74) vs pH 8.7 (W87)). Next, to identify which metabolites contributed to the separation, 2-way orthogonal comparisons were made between each cell line with the two different growth conditions (Figure 9B,C). Key metabolites contributing to the separation were selected by VIP values higher than one from all 2-way comparisons. Additionally, the selected metabolites were cross-checked by univariate t-test analysis (with the false discovery rate Q ≤ 0.05). Three metabolites are changed significantly more in Δak cells than in WT, all increased in pH 8.7 compared with pH 7.4, and 15 are changed significantly more in WT cells than in Δak, all decreased (see Supplementary Tables S2 and S3). The role of these metabolites in the response to high pH and their interactions with AK remains to be investigated.
Table 2. Cell counts after 2 h of growth in medium pH 7.4 or 8.7.
| Cell type | pH 7.4 | pH 8.7 | Decrease (%) |
|---|---|---|---|
| 427 WT/ml (×106) | 6.27 ± 0.1 | 5.54 ± 0.47 | 22 |
| Δak ml−1 (×106) | 5.79 ± 0.25 | 3.37 ± 0.12 | 52 |
Figure 9. OPLS-DA scores plot of the metabolomics pH experiment.
(A) OPLS-DA including all four groups: WT and Δak cells at pH 7.4 and 8.7. (B) OPLS-DA including only Δak cells at both pH. (C) OPLS-DA including only WT cells at both pH.
Unexpected metabolites
Using isotope labelled substrates allows detection of unexpected and novel metabolites too. For example, tracing U-13C glucose showed the presence of high carbon sugar phosphates such as octulose 8-phosphate and nonulose 9-phosphate as well as a number of pyruvate-derived conjugates in T. brucei [5]. Using U-13C amino acids in the experiments reported here we are able to add information on the provenance of some of the metabolites identified in the glucose labelling experiments as well as detect several new unknown metabolites (see Table 3). Creek et al. detected metabolites with masses corresponding to carboxyethyl-l-arginine and carboxyethyl-l-ornithine with 3 carbons 13C labels derived from glucose. Labelled arginine confirmed the presence of these two metabolites with the 6 non-glucose derived carbons of ‘carboxyethyl-l-arginine’ (or d-octopine) coming from arginine and the 5 non-glucose carbons of ‘carboxyethyl-l-ornithine’ coming from arginine via ornithine.
Table 3. New metabolites.
| Formula | Carbon labelled | Putative source |
|---|---|---|
| C8H16N2O4 | 5 from arginine, 3 from glucose | l-Ornithine + pyruvate? |
| C9H18N4O4 | 6 from arginine, 3 from glucose | l-Arginine + pyruvate? |
| C8H11NO6S | 5 from glutamine, 3 from cysteine | Oxoglutarate + cysteine? Product of an aminotransferase reaction using glutamylcysteine as a substrate? |
| C10H14N2O7S | 5 from glutamine, 3 from cysteine | C8H11NO6S + glycine? Product of an aminotransferase reaction using glutathion as a substrate? |
| C6H11NO4S | 3 from cysteine, 3 from glucose | Cysteine + pyruvate? |
| C6H9NO4S | 3 from cysteine, 3 from glucose | C6H11NO4S–H2? |
| C8H17NO6S | 3 from cysteine, 5 from glucose | Cysteine + pentose? |
The formula are assigned based on a measured m/z within 3 ppm of the calculated m/z. No putative identification could be made that would match mass and labelling patterns.
The metabolite with a formula C6H9NO4S was hypothesised to be a pyruvate-cysteine adduct [5] and the cysteine labelling experiment confirms that the 3 non-glucose derived carbons are from cysteine. A related metabolite of formula C6H11NO4S which has a different retention time and the same carbon labelling pattern was also found. A third metabolite containing labelled carbon derived from both glucose and cysteine of formula C8H17NO6S has a labelling pattern suggestive of its being formed from cysteine and a pentose, most likely ribose. Roles for these metabolites have not been assigned, and they have not been described in other systems.
Finally, we can also see two metabolites formed from 5 carbons coming from glutamine and 3 carbons from cysteine. One has a mass that matches the formula C8H11NO6S and could be produced from 2-oxoglutarate and cysteine, possibly the same way glutamylcysteine is produced (see Figure 2). The other has a mass that matches the formula C10H14N2O7S which could be C8H11NO6S used by glutathione synthetase instead of glutamylcysteine.
All these unexpected metabolites are absent from the fresh medium samples. They could be produced either by non-enzymatic reactions caused by the accumulation of particular metabolites within cells, or by enzymes using non-conventional substrates, as is the case for hydroxyglutarate (see section Glutamine). In culture, pyruvate accumulates to high concentrations inside and outside of the cells [18,35] which could explain the pyruvate derived metabolites. However, for the glutamine and cysteine derived metabolites accidental enzymatic reactions seem more likely.
Conclusion
Here we showed how T. brucei bloodstream form cells metabolise five amino acids: glutamine, cysteine, proline, methionine and arginine. In addition to confirming the operation of a range of pathways, both catabolic and anabolic, we detected possible new metabolites, roles for which will need to be further investigated. Our results also indicate that, as shown in previous proteomics datasets [63], pathways are likely present in the mitochondrion that were previously ignored or downplayed, particularly for glutamine consumption. However, only a very low amount of proline is used by the parasite, indicating that the enzymes are present but only minimally active, perhaps allowing the cells to be more resistant to changes in their environment.
Supporting information
Supplementary Figures.
Acknowledgments
We thank Dr Federica Giordani for her help collecting the arginine labelled cell free medium samples. The mzXML files of all the experiments described in this study are available in the Metabolights database at https://www.ebi.ac.uk/metabolights/MTBLS624.
Abbreviations
- AdoMet
S-adenosylmethionine
- AK
arginine kinase
- CMM
Creek’s ‘Minimal’ Media
- LC–MS
liquid chromatography–mass spectrometry
- MTA
methyl-5′-thioadenosine
- MTR
methylthioribose
- NOS
nitric oxide synthase
- OPLS-DA
orthogonal partial least squares-discriminant analysis
- VIP
variable importance in projection values
Funding
This work was supported by the Leverhulme Trust [grant number ECF-2015-392 (to FA)]; the Wellcome Trust core grant to the Wellcome Centre for Integrative Parasitology [grant number 104111/Z/14/Z (to MPB)]; and the Doctoral Training Centre (DTC) in Technologies at the Interface between Engineering, the Physical Sciences and the Life Sciences, University of [grant number EP/F500424/1 (to KJ)]. The DTC is funded by the Engineering and Physical Sciences Research Council (EPSRC) and the Biotechnology and Biological Sciences Research Council (BBSRC). The mzXML files of all the experiments described in the present study are available in the Metabolights database at https://www.ebi.ac.uk/metabolights/MTBLS624.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Conceptualisation: FA, MBP; data curation: FA, DHK, KJ; formal analysis: FA, DHK; funding acquisition: FA, RB, MPB; investigation: KJ, DHK, EJK; resources: RB, MPB; supervision: FA, RB, MPB; visualisation: FA, DHK; writing – original draft: FA; writing – review and editing: FA, MPB, DHK, EJK, RB, KJ.
References
- 1.Bienen E.J., Hammadi E. and Hill G.C. (1981) Trypanosoma brucei: biochemical and morphological changes during in vitro transformation of bloodstream to procyclic-trypomastigotes. Exp. Parasitol. 51, 408–417 10.1016/0014-4894(81)90128-4 [DOI] [PubMed] [Google Scholar]
- 2.Hart D.T., Misset O., Edwards S.W. and Opperdoes F.R. (1984) A comparison of the glycosomes (microbodies) isolated from Trypanosoma brucei bloodstream form and cultured procyclic trypomastigotes. Mol. Biochem. Parasitol. 12, 25–35 10.1016/0166-6851(84)90041-0 [DOI] [PubMed] [Google Scholar]
- 3.Bringaud F., Rivière L. and Coustou V. (2006) Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 149, 1–9 10.1016/j.molbiopara.2006.03.017 [DOI] [PubMed] [Google Scholar]
- 4.Chokkathukalam A., Jankevics A., Creek D.J., Achcar F., Barrett M.P. and Breitling R. (2013) mzMatch-ISO: an R tool for the annotation and relative quantification of isotope-labelled mass spectrometry data. Bioinformatics 29, 281–283 10.1093/bioinformatics/bts674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creek D.J., Mazet M., Achcar F., Anderson J., Kim D.-H., Kamour R.. et al. (2015) Probing the metabolic network in bloodstream-form Trypanosoma brucei using untargeted metabolomics with stable isotope labelled glucose. PLoS Pathog. 11, e1004689 10.1371/journal.ppat.1004689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creek D., Nijagal B., Kim D., Rojas F., Matthews K. and Barrett M. (2013) Metabolomics guides rational development of a simplified cell culture medium for drug screening against Trypanosoma brucei. Antimicrob. Agents Chemother. 57, 2768–2779 10.1128/AAC.00044-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazet M., Morand P., Biran M., Bouyssou G., Courtois P., Daulouède S.. et al. (2013) Revisiting the central metabolism of the bloodstream forms of Trypanosoma brucei: production of acetate in the mitochondrion is essential for parasite viability. PLoS Negl. Trop. Dis. 7, e2587 10.1371/journal.pntd.0002587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGettrick A.F., Corcoran S.E., Barry P.J.G., McFarland J., Crès C., Curtis A.M.. et al. (2016) Trypanosoma brucei metabolite indolepyruvate decreases HIF-1α and glycolysis in macrophages as a mechanism of innate immune evasion. Proc. Natl. Acad. Sci. U.S.A. 113, E7778–E7787 10.1073/pnas.1608221113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cross M., Kieft R., Sabatini R., Dirks-Mulder A., Chaves I. and Borst P. (2002) J-binding protein increases the level and retention of the unusual base J in trypanosome DNA. Mol. Microbiol. 46, 37–47 10.1046/j.1365-2958.2002.03144.x [DOI] [PubMed] [Google Scholar]
- 10.Begolo D., Erben E. and Clayton C. (2014) Drug target identification using a trypanosome overexpression library. Antimicrob. Agents Chemother. 58, 6260–6264 10.1128/AAC.03338-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J. and Russell D.W. (2001) Preparation and analysis of eukaryotic genomic DNA. In Molecular Cloning: A Laboratory Manual, vol. 6, pp. 6.39–6.58, CSHL Press [Google Scholar]
- 12.Adusumilli R. and Mallick P. (2017) Data conversion with ProteoWizard msConvert. Methods Mol. Biol. 1550, 339–368 10.1007/978-1-4939-6747-6_23 [DOI] [PubMed] [Google Scholar]
- 13.Benton H.P., Wong D.M., Trauger S.A. and Siuzdak G. (2008) XCMS2: processing tandem mass spectrometry data for metabolite identification and structural characterization. Anal. Chem. 80, 6382–6389 10.1021/ac800795f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheltema R.A., Jankevics A., Jansen R.C., Swertz M.A. and Breitling R. (2011) PeakML/mzMatch: a file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 83, 2786–2793 10.1021/ac2000994 [DOI] [PubMed] [Google Scholar]
- 15.Sumner L.W., Lei Z., Nikolau B.J., Saito K., Roessner U. and Trengove R. (2014) Proposed quantitative and alphanumeric metabolite identification metrics. Metabolomics 10, 1047–1049 10.1007/s11306-014-0739-6 [DOI] [Google Scholar]
- 16.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A.. et al. (2007) Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia J., Sinelnikov I.V., Han B. and Wishart D.S. (2015) MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D.-H., Achcar F., Breitling R., Burgess K.E. and Barrett M.P. (2015) LC-MS-based absolute metabolite quantification: application to metabolic flux measurement in trypanosomes. Metabolomics 11, 1721–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M.. et al. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457–62 10.1093/nar/gkp851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., Leija C., Rijo-Ferreira F., Chen J., Cestari I., Stuart K.. et al. (2015) GMP synthase is essential for viability and infectivity of Trypanosoma brucei despite a redundant purine salvage pathway. Mol. Microbiol. 97, 1006–1020 10.1111/mmi.13083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nara T., Gao G., Yamasaki H., Nakajima-Shimada J. and Aoki T. (1998) Carbamoyl-phosphate synthetase II in kinetoplastids. Biochim. Biophys. Acta 1387, 462–468 10.1016/S0167-4838(98)00127-7 [DOI] [PubMed] [Google Scholar]
- 22.Hammond D.J. and Gutteridge W.E. (1982) UMP synthesis in the kinetoplastida. Biochim. Biophys. Acta 718, 1–10 10.1016/0304-4165(82)90002-2 [DOI] [PubMed] [Google Scholar]
- 23.Alsford S., Turner D.J., Obado S.O., Sanchez-Flores A., Glover L., Berriman M.. et al. (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21, 915–924 10.1101/gr.115089.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loureiro I., Faria J., Clayton C., Ribeiro S.M., Roy N., Santarém N.. et al. (2013) Knockdown of asparagine synthetase A renders Trypanosoma brucei auxotrophic to asparagine. PLoS Negl. Trop. Dis. 7, e2578 10.1371/journal.pntd.0002578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markos A., Blahůsková A., Kalous M., Bysková E., Byska K. and Nohýnková E. (1989) Metabolic differentiation of bloodstream forms of Trypanosoma brucei brucei into procyclic forms. Effect of hydroxyurea, arabinosyl adenine, and serum omission. Folia Parasitol. (Praha) 36, 225–238 [PubMed] [Google Scholar]
- 26.Marciano D., Llorente C., Maugeri D.A., de la Fuente C., Opperdoes F., Cazzulo J.J.. et al. (2008) Biochemical characterization of stage-specific isoforms of aspartate aminotransferases from Trypanosoma cruzi and Trypanosoma brucei. Mol. Biochem. Parasitol. 161, 12–20 10.1016/j.molbiopara.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 27.Spitznagel D., Ebikeme C., Biran M., Nic a′ Bháird N., Bringaud F., Henehan G.T.M.. et al. (2009) Alanine aminotransferase of Trypanosoma brucei – a key role in proline metabolism in procyclic life forms. FEBS J. 276, 7187–7199 10.1111/j.1742-4658.2009.07432.x [DOI] [PubMed] [Google Scholar]
- 28.Marciano D., Maugeri D.A., Cazzulo J.J. and Nowicki C. (2009) Functional characterization of stage-specific aminotransferases from trypanosomatids. Mol. Biochem. Parasitol. 166, 172–182 10.1016/j.molbiopara.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 29.Durieux P.O., Schütz P., Brun R. and Köhler P. (1991) Alterations in Krebs cycle enzyme activities and carbohydrate catabolism in two strains of Trypanosoma brucei during in vitro differentiation of their bloodstream to procyclic stages. Mol. Biochem. Parasitol. 45, 19–27 10.1016/0166-6851(91)90023-Y [DOI] [PubMed] [Google Scholar]
- 30.Rivière L., van Weelden S.W.H., Glass P., Vegh P., Coustou V., Biran M.. et al. (2004) Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism. J. Biol. Chem. 279, 45337–45346 10.1074/jbc.M407513200 [DOI] [PubMed] [Google Scholar]
- 31.Vernal J., Muñoz-Jordán J., Müller M., Cazzulo J.J. and Nowicki C. (2001) Sequencing and heterologous expression of a cytosolic-type malate dehydrogenase of Trypanosoma brucei. Mol. Biochem. Parasitol. 117, 217–221 10.1016/S0166-6851(01)00343-7 [DOI] [PubMed] [Google Scholar]
- 32.Aranda A., Maugeri D., Uttaro A.D., Opperdoes F., Cazzulo J.J. and Nowicki C. (2006) The malate dehydrogenase isoforms from Trypanosoma brucei: subcellular localization and differential expression in bloodstream and procyclic forms. Int. J. Parasitol. 36, 295–307 10.1016/j.ijpara.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 33.Tait A., Barry J.D., Wink R., Sanderson A. and Crowe J.S. (1985) Enzyme variation in T. brucei ssp. II. Evidence for T. b. rhodesiense being a set of variants of T. b. brucei. Parasitology 90(Pt 1), 89–100 10.1017/S0031182000049040 [DOI] [PubMed] [Google Scholar]
- 34.Intlekofer A.M., Wang B., Liu H., Shah H., Carmona-Fontaine C., Rustenburg A.S.. et al. (2017) l-2-Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat. Chem. Biol. 13, 494–500 10.1038/nchembio.2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiemer E.A., Ter Kuile B.H., Michels P.A. and Opperdoes F.R. (1992) Pyruvate transport across the plasma membrane of the bloodstream form of Trypanosoma brucei is mediated by a facilitated diffusion carrier. Biochem. Biophys. Res. Commun. 184, 1028–1034 10.1016/0006-291X(92)90694-G [DOI] [PubMed] [Google Scholar]
- 36.Tait A., Babiker E.A. and Le Ray D. (1984) Enzyme variation in Trypanosoma brucei spp. I. Evidence for the sub-speciation of Trypanosoma brucei gambiense. Parasitology 89, 311–326 10.1017/S0031182000001335 [DOI] [PubMed] [Google Scholar]
- 37.Leroux A.E., Maugeri D.A., Opperdoes F.R., Cazzulo J.J. and Nowicki C. (2011) Comparative studies on the biochemical properties of the malic enzymes from Trypanosoma cruzi and Trypanosoma brucei. FEMS Microbiol. Lett. 314, 25–33 10.1111/j.1574-6968.2010.02142.x [DOI] [PubMed] [Google Scholar]
- 38.Dean S., Marchetti R., Kirk K. and Matthews K.R. (2009) A surface transporter family conveys the trypanosome differentiation signal. Nature 459, 213–217 10.1038/nature07997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saas J., Ziegelbauer K., von Haeseler A., Fast B. and Boshart M. (2000) A developmentally regulated aconitase related to iron-regulatory protein-1 is localized in the cytoplasm and in the mitochondrion of Trypanosoma brucei. J. Biol. Chem. 275, 2745–2755 10.1074/jbc.275.4.2745 [DOI] [PubMed] [Google Scholar]
- 40.Mantilla B.S., Marchese L., Casas-Sánchez A., Dyer N.A., Ejeh N., Biran M.. et al. (2017) Proline metabolism is essential for Trypanosoma brucei brucei survival in the tsetse vector. PLoS Pathog. 13, e1006158 10.1371/journal.ppat.1006158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamour N., Rivière L., Coustou V., Coombs G.H., Barrett M.P. and Bringaud F. (2005) Proline metabolism in procyclic Trypanosoma brucei is down-regulated in the presence of glucose. J. Biol. Chem. 280, 11902–11910 10.1074/jbc.M414274200 [DOI] [PubMed] [Google Scholar]
- 42.Berger L.C., Wilson J., Wood P. and Berger B.J. (2001) Methionine regeneration and aspartate aminotransferase in parasitic protozoa. J. Bacteriol. 183, 4421–4434 10.1128/JB.183.15.4421-4434.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christa L., Kersual J., Augé J. and Pérignon J.L. (1988) Methylthioadenosine toxicity and metabolism to methionine in mammalian cells. Biochem. J. 255, 145–152 10.1042/bj2550145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bacchi C., Sanabria K., Spiess A., Vargas M., Marasco C., Jimenez L.. et al. (1997) In vivo efficacies of 5′-methylthioadenosine analogs as trypanocides. Antimicrob. Agents Chemother. 41, 2108–2112 10.1128/AAC.41.10.2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghoda L.Y., Savarese T.M., Northup C.H., Parks R.E., Garofalo J., Katz L.. et al. (1988) Substrate specificities of 5′-deoxy-5′-methylthioadenosine phosphorylase from Trypanosoma brucei brucei and mammalian cells. Mol. Biochem. Parasitol. 27, 109–118 10.1016/0166-6851(88)90030-8 [DOI] [PubMed] [Google Scholar]
- 46.Hughes J.A. (2006) In vivo hydrolysis of S-adenosyl-l-methionine in Escherichia coli increases export of 5-methylthioribose. Can. J. Microbiol. 52, 599–602 10.1139/w06-008 [DOI] [PubMed] [Google Scholar]
- 47.Henderson D.M., Hanson S., Allen T., Wilson K., Coulter-Karis D.E., Greenberg M.L.. et al. (1992) Cloning of the gene encoding Leishmania donovani S-adenosylhomocysteine hydrolase, a potential target for antiparasitic chemotherapy. Mol. Biochem. Parasitol. 53, 169–183 10.1016/0166-6851(92)90019-G [DOI] [PubMed] [Google Scholar]
- 48.Parker N.B., Yang X., Hanke J., Mason K.A., Schowen R.L., Borchardt R.T.. et al. (2003) Trypanosoma cruzi: molecular cloning and characterization of the S-adenosylhomocysteine hydrolase. Exp. Parasitol. 105, 149–158 10.1016/j.exppara.2003.10.001 [DOI] [PubMed] [Google Scholar]
- 49.Nozaki T., Shigeta Y., Saito-Nakano Y., Imada M. and Kruger W.D. (2001) Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine beta-synthase and serine acetyltransferase from Trypanosoma. J. Biol. Chem. 276, 6516–6523 10.1074/jbc.M009774200 [DOI] [PubMed] [Google Scholar]
- 50.Giordana L., Mantilla B.S., Santana M., Silber A.M. and Nowicki C. (2014) Cystathionine γ-lyase, an enzyme related to the reverse transsulfuration pathway, is functional in Leishmania spp. J. Eukaryot. Microbiol. 61, 204–213 10.1111/jeu.12100 [DOI] [PubMed] [Google Scholar]
- 51.Bacchi C.J., Goldberg B., Garofalo-Hannan J., Rattendi D., Lyte P. and Yarlett N. (1995) Fate of soluble methionine in African trypanosomes: effects of metabolic inhibitors. Biochem. J. 309, 737–743 10.1042/bj3090737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duszenko M., Mühlstädt K. and Broder A. (1992) Cysteine is an essential growth factor for Trypanosoma brucei bloodstream forms. Mol. Biochem. Parasitol. 50, 269–273 10.1016/0166-6851(92)90224-8 [DOI] [PubMed] [Google Scholar]
- 53.Opperdoes F.R. and Michels P.A.M. (2007) Horizontal gene transfer in trypanosomatids. Trends Parasitol. 23, 470–476 10.1016/j.pt.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 54.Yarlett N. and Bacchi C.J. (1988) Effect of dl-alpha-difluoromethylornithine on methionine cycle intermediates in Trypanosoma brucei brucei. Mol. Biochem. Parasitol. 27, 1–10 10.1016/0166-6851(88)90019-9 [DOI] [PubMed] [Google Scholar]
- 55.Rivière L., Moreau P., Allmann S., Hahn M., Biran M., Plazolles N.. et al. (2009) Acetate produced in the mitochondrion is the essential precursor for lipid biosynthesis in procyclic trypanosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 12694–12699 10.1073/pnas.0903355106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C.. et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- 57.Hai Y., Kerkhoven E.J., Barrett M.P. and Christianson D.W. (2015) Crystal structure of an arginase-like protein from Trypanosoma brucei that evolved without a binuclear manganese cluster. Biochemistry 54, 458–471 10.1021/bi501366a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent I., Creek D., Burgess K., Woods D., Burchmore R. and Barrett M. (2012) Untargeted metabolomics reveals a lack of synergy between nifurtimox and eflornithine against Trypanosoma brucei. PLoS Negl. Trop. Dis. 6, e1618 10.1371/journal.pntd.0001618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sternberg M.J. and Mabbott N.A. (1996) Nitric oxide-mediated suppression of T cell responses during Trypanosoma brucei infection: soluble trypanosome products and interferon-gamma are synergistic inducers of nitric oxide synthase. Eur. J. Immunol. 26, 539–543 10.1002/eji.1830260306 [DOI] [PubMed] [Google Scholar]
- 60.Pereira C.A., Alonso G.D., Torres H.N. and Flawiá M.M. (2002) Arginine kinase: a common feature for management of energy reserves in African and American flagellated trypanosomatids. J. Eukaryot. Microbiol. 49, 82–85 10.1111/j.1550-7408.2002.tb00346.x [DOI] [PubMed] [Google Scholar]
- 61.Alonso G.D., Pereira C.A., Remedi M.S., Paveto M.C., Cochella L., Ivaldi M.S.. et al. (2001) Arginine kinase of the flagellate protozoa Trypanosoma cruzi. Regulation of its expression and catalytic activity. FEBS Lett. 498, 22–25 10.1016/S0014-5793(01)02473-5 [DOI] [PubMed] [Google Scholar]
- 62.Pereira C.A., Alonso G.D., Ivaldi S., Silber A.M., Alves M.J.M., Torres H.N.. et al. (2003) Arginine kinase overexpression improves Trypanosoma cruzi survival capability. FEBS Lett. 554, 201–205 10.1016/S0014-5793(03)01171-2 [DOI] [PubMed] [Google Scholar]
- 63.Zíková A., Verner Z., Nenarokova A., Michels P.A.M. and Lukeš J. (2017) A paradigm shift: the mitoproteomes of procyclic and bloodstream Trypanosoma brucei are comparably complex. PLoS Pathog. 13, e1006679 10.1371/journal.ppat.1006679 [DOI] [PMC free article] [PubMed] [Google Scholar]