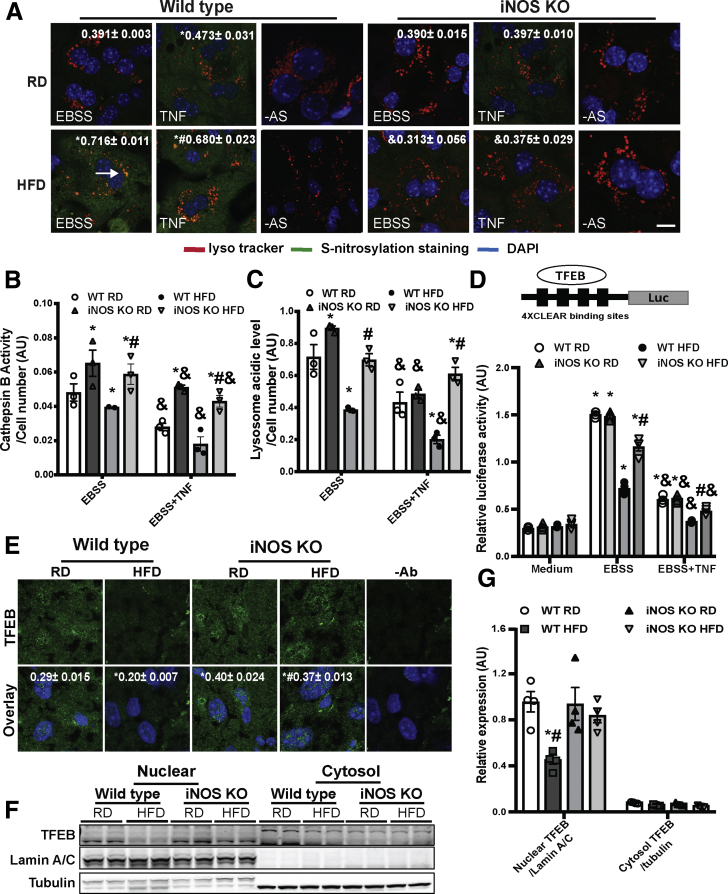
Figure 3.
iNOS induces hepatic lysosomal nitrosative stress. (A) Representative confocal images (40×) of S-nitrosylated proteins in primary hepatocytes from WT and iNOS-KO mice (n = 3) using a modified in situ biotin-switch method. Autophagy was induced by EBSS (4 hours) and cells were pretreated with TNF (10 ng/mL, 16 hours). –AS: no ascorbate as a negative control for biotin-switch assay. Scale bar: 10 μm. Quantification of the S-nitrosylated protein and lysotracker are shown on the top of each image. Data are shown as mean ± SEM as determined by Person’s correlation coefficient. * indicates statistically significant difference relative to EBSS in WT RD, and # indicates statistical significance between HFD group determined by analysis of variance (ANOVA) followed by post hoc test (P < .05; 10 fields/group; n = 3, biological replicates; 12 weeks on HFD). (B) CTSB activity and (C) lysosomal acidity in live primary hepatocytes from WT and iNOS-KO mice. * indicates statistical significance compared with WT RD in the same treatment, # indicates statistical significance within HFD groups in the same treatment, and & indicates statistical significance between EBSS- and TNF-treated groups in the same cell type, determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). (D) TFEB activity in primary hepatocytes from WT and iNOS-KO mice. Cells were transfected with the indicated constructs for 48 hours, and treated with EBSS (4 hours) ± TNF (10 ng/mL, 16 hours). The data were normalized to Renilla luciferase. * indicates statistical significance compared with WT RD medium from the same cell type, # indicates statistical significance between HFD groups in the same treatment, and & indicates statistical significance between EBSS and TNF in the same cell type determined by ANOVA followed by a post hoc test (P < .05; n = 3, biological replicates; 12 weeks on HFD). The 4XCLEAR construct is shown on the top of the panel. (E) Representative confocal images (63×) for TFEB staining in livers of WT mice and iNOS KO mice. Green: TFEB; blue: DAPI. –Ab: nonantibody controls. Scale bar: 10 μm. Quantified colocalization of TFEB with DAPI are shown at the top of each image. Data are shown as Pearson’s correlation coefficient as mean ± SEM. * indicates statistically significant difference relative to WT RD, and # indicates statistical significance between HFD group determined by ANOVA followed by a post hoc test (P < .05; 10 fields/group; n = 3, biological replicates; 16 weeks on HFD). (F) Representative Western blot analysis of TFEB nuclear translocation in panel E. Each sample is the combined fraction from livers of 6 mice. (G) Densitometry of Western blot analysis in panel F. * indicates statistical significance between HFD and RD in same type of mouse, # indicates statistical significance between HFD group, determined by ANOVA followed by post hoc test (P < .05; each lane is the combined fraction from livers of 3 mice; n = 12, biological replicates).