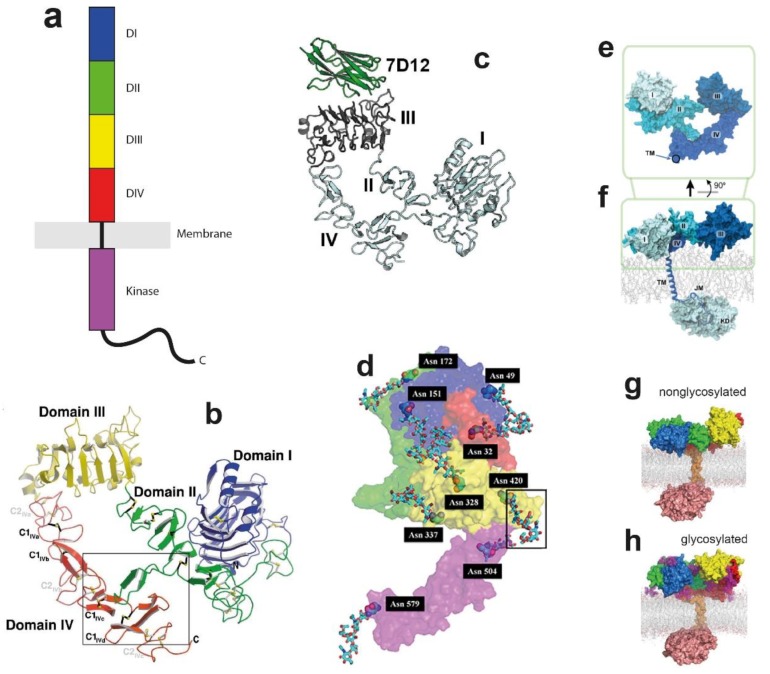
Figure 1.
Topology, structure, and simulations of the epidermal growth factor receptor (EGFR) monomer. (a) Cartoon of an EGFR monomer, showing its domain structure. (b) Ribbon representation of the solubilised monomeric extracellular module (ECM) of EGFR (sEGFR) structure (Protein Data Bank (PDB): 1NQL), showing the “tether”, in which an α-hairpin from DII forms an interaction with DIV. Disulfide bonds are labelled in black (C1) and gray (C2). Taken, with permission, from Ferguson et al. [85]. (c) Ribbon representation of sEGFR with bound VHH domain 7D12 (PDB: 4KRL), again showing the presence of the tether. Taken, with permission, from Schmitz et al. [89]]. (d) Extended monomer structure of EGFR (PDB: 3NJP), taking a monomer unit from the ligand-bound back-to-back dimer. The locations of glycosylated Asparagine residues are shown, one highlighted inside a grey box. Taken, with permission, from Irani [93]. (e) A molecular dynamic (MD) simulation of the sEGFR started from the structure in d. The connecting point between the extracellular and the transmembrane helices is marked by a circle. (f) As (e) but showing the near-complete receptor. Taken, with permission, from Arkhipov et al. [67]. (g,h) Endpoint structures from 1 µs simulations of the (g) nonglycosylated and (h) glycosylated EGFR in 1,2-dioleoylsn-glycero-3-phosphocholine (DOPC)/ sphingomyelin (SM)/cholesterol membranes. Subdomain DI shown in blue, DII in green, DIII in yellow and DIV in red. The single-pass transmembrane (TM) domain is shown in orange, the intracellular tyrosine kinase domain (TKD) in salmon, and the glycans in purple. Taken, with permission, from Kaszuba et al. [94].