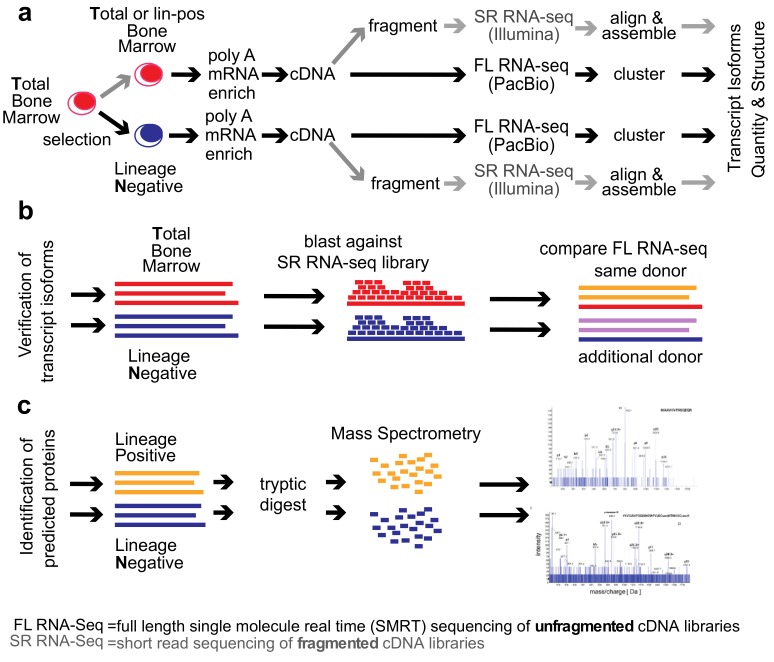
Figure 1.
Graphical abstract of the Transcriptome analysis of human bone marrow (BM) cell populations. (a) Flow of experiments and analyses. Poly(A)+ RNA was isolated from Total (T, red) or lineage-negative BM cell populations (N, blue). Unfragmented, full-length complementary DNA (cDNA) libraries were subjected to single-molecule real-time (SMRT) RNA-seq (PacBio platform) or conventional short-read RNA-seq of fragmented cDNAs at 20 million (T) or 100 million (N) read depth (Illumina). Full-length RNA-seq data were processed using the ToFU platform. Illumina reads were first aligned and then assembled using the Tuxedo suite. The efficiency of the double selection for lineage-negative cells used here was confirmed by comparison of the abundance of standard markers of differentiated cells: CD14 = 6:1; CD16b = 25:1; CD24 = 109:1; CD45 = 11:1; CD66b = 16:1 expression ratio of lineage-positive to lineage-negative cells. (b) Validation of transcript isoforms identified by full-length RNA-seq. A BLAST-able library was generated from the short-read RNA-seq and used to align the isoforms identified by full-length RNA-seq. A separate full-length RNA-seq of an independent sample from the same donor, and a sample from a different donor were used for comparison. (c) Identification of proteins predicted by full-length RNA-seq using mass spectrometry of bone marrow cell extracts.