Figure 3.
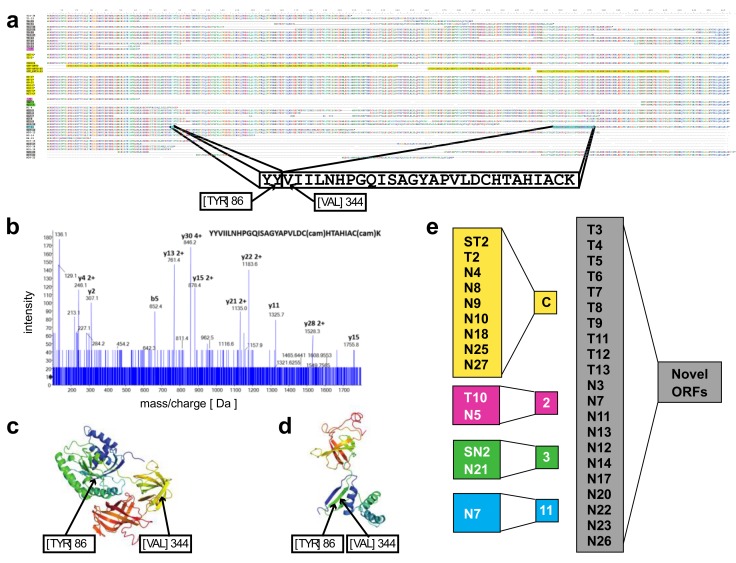
Proteins predicted from the transcript isoforms identified for EEF1A1. (a) Amino acid sequence alignments including the transcript isoform identifiers (see Figure 2b,c). The protein predicted from the N7-11 transcript isoform (highlighted in blue) joins Y86 and V344 in the canonical protein. The amino acid sequence of a predicted signature tryptic fragment peptide is shown. (b) Manually assigned fragmentation spectra from targeted analysis by Nano LC-MS/MS of proteins extracted from lineage-negative bone marrow cells with the sequence read of the N7-24 signature peptide; cam, carbamidomethylation. (c,d) Protein structure models were generated in Phyre2 [23]. The predicted structure of the canonical EEF1A1 protein P68104 (c) and of the novel N7 protein (d) are shown. The c1g7ca template of P68104 used for the model covered 90% of the amino acids in the N7 isoform. (e) Higher magnification of transcript isoform identifiers that code for the canonical protein (yellow background). C, canonical RefSeq derived transcripts. C*, previously unknown transcript isoforms that code for the canonical protein. Transcript isoforms SN2-3 and N21-3 predict the same protein (green background).