Figure 1.
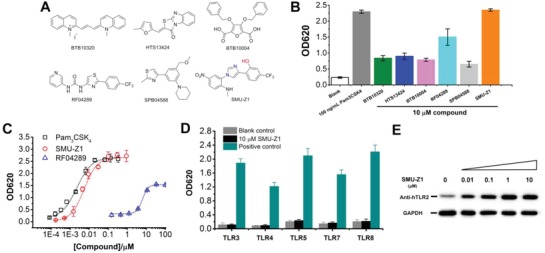
Structures of the hit compounds obtained from HTS, and bioactivity validation of the optimized compound SMU‐Z1. A) Chemical structures of the hits from the HTS. B) The initial hit compounds obtained for TLR2 activation. HEK‐Blue hTLR2 cells were incubated with indicated compounds for 24 h, and the cell culture supernatants were detected by QUANTI‐Blue (Invivogen) to evaluate SEAP signaling at 620 nm (OD620). Pam3CSK4 was used as a positive control in the experiment. C) Comparison of SMU‐Z1 with RF04289 and Pam3CSK4 for the SEAP activation in HEK‐Blue hTLR2 cells. D) HEK‐Blue human TLR3, TLR4, TLR5, TLR7, and TLR8 cells were incubated with SMU‐Z1 (10 × 10−6 m) with the control of TLR‐specific agonists separately for 24 h, and the activation was evaluated by QUANTI‐Blue at OD620. As positive control, agonists that selectively activate a specific TLR were used: TLR3, Poly I:C (10 µg mL−1); TLR4, LPS (10 ng mL−1); TLR5, FLA‐BS (1 µg mL−1); TLR7 and TLR8, R848 (5 µg mL−1). E) SMU‐Z1 stabilized more TLR2 protein in HEK‐Blue hTLR2 cells as the concentrations increase in the cultures. Cells were treated with SMU‐Z1 (0–10 × 10−6 m) for 24 h, and the cell lysates were detected using Western blot and rabbit anti‐TLR2 antibody. GAPDH served as the cell lysates input control. Data presented are mean ± SD and the figures shown are representative of three independent experiments.
