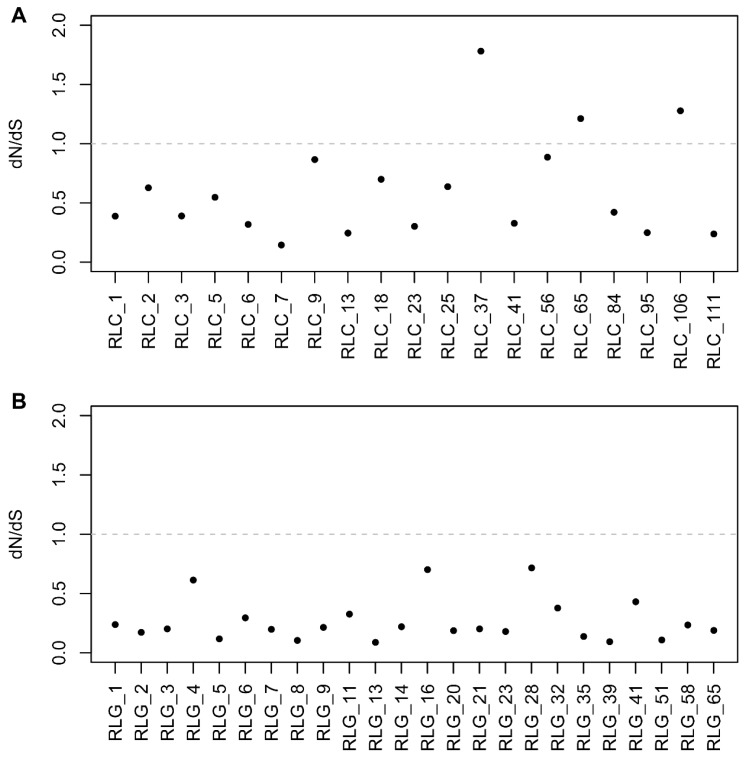
Figure 7.
Nonsynonymous/Synonymous (dN/dS) rates for the LTR retrotransposons. (A) dN/dS rates for the Copia elements; (B) dN/dS rates for the Gypsy elements. The x-axis represents different families. The y-axis displays values of dN/dS. Only full-length sequences with intact RT genes were retained for selective pressure analysis. Protein sequences of RT genes were aligned by MUSCLE, then PAN2NAL was utilized to convert the protein MSA (multiple sequence alignment) format to a DNA codon-based alignment with the universal code model. The codeml module was used to perform dN/dS calculations. The rates of dN/dS reflected the selective pressures of these elements, dN/dS < 1, dN/dS = 1, and dN/dS > 1 denote purifying selection, neutral mutations, and adaptive molecular evolution, respectively.