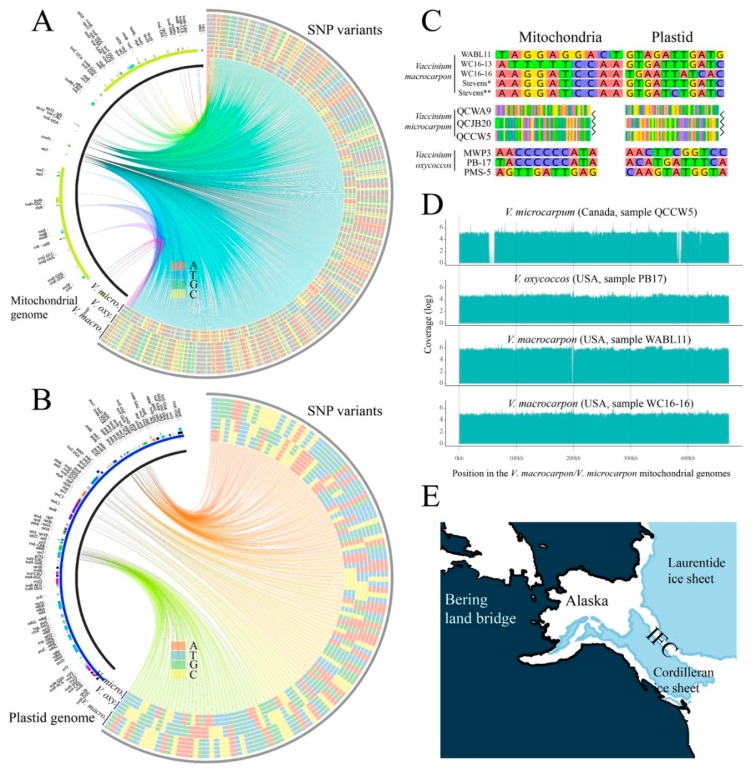
Figure 4.
SNP variants among V. macrocarpon, V. microcarpum, and V. oxycoccos. SNP (single nucleotide polymorphism) variants detected in mitochondria (A) and plastid (B) genomes among all 11 samples sequenced with both Illumina and PacBio. On the heatmaps (right arcs), the inner three rings correspond to the V. microcarpum samples QCCW5, QCJB20, and QCWA9; the next three rings (4–6) correspond to the V. oxycoccos samples MWPB3, PB17, and PMS5; rings 7–9 correspond to the wild V. macrocarpon samples WABL11, WC16-13, and WC16-16; and rings 10–11 correspond to cultivated V. macrocarpon sequenced with Illumina and PacBio, respectively (note these last two sequences were identical to the Alaskan V. microcarpum sequences). Each SNP is connected by a colored link to its physical position within either the mitochondria or plastid genome (left to right arcs); link colors are meaningless. For reference purposes, gene locations were added on the left arcs and colored by category. (C) SNP variants were called when aligning sequences by species; for the V. macrocarpon alignment, Stevens* correspond to the assembly carried out with Illumina data (used to validate the PacBio assembly), whereas Stevens** correspond to the one with PacBio; for V. microcarpum, only the first positions of the alignment are shown. (D) Coverage plots using Illumina filtered raw reads and V. macrocarpon/V. microcarpum mitochondrial genomes as reference (additional coverage plots are provided in Figures S1 and S2). (E) Ice extent between 9–10 k years ago showing the ice-free corridor (IFC).