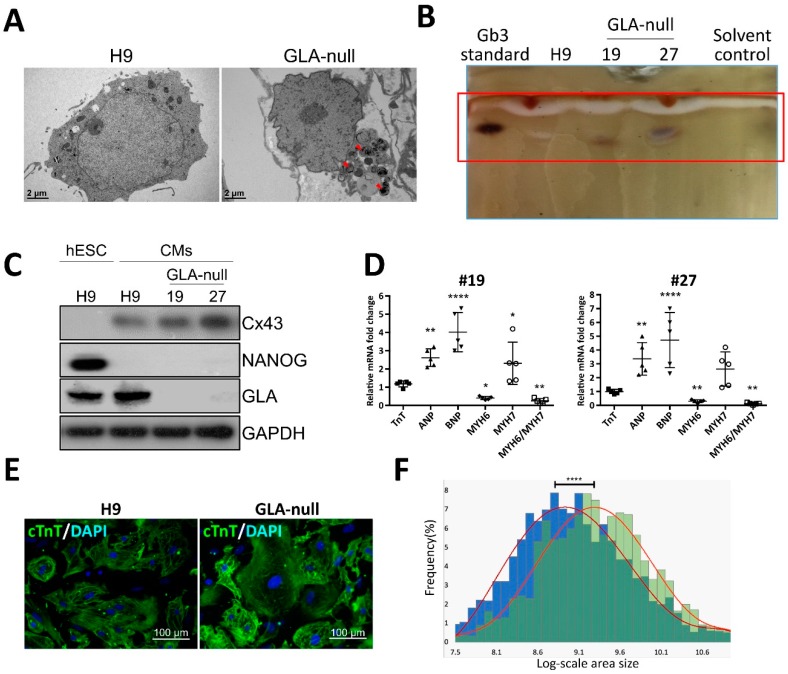
Figure 3.
Recapitulation of FD-specific cardiac abnormalities in GLA-null cardiomyocytes. (A) TEM images showing the ultrastructure of parental type (H9) and GLA-null CMs. The red arrowheads indicate the multilayered lysosomal structure. (B) Gb3 content in parental (H9) and GLA-null CMs (clones #19 and #27) analyzed by TLC. (C) Western blot showing lack of expression of GLA in clones #19 and #27 of GLA-null CMs as compared to the parental wild type CMs and hESCs (H9). Connexin 43 (Cx43) and NANOG served as cardiomyocyte and pluripotency markers, respectively. GAPDH used as a loading control. (D) qRT-PCR analysis of expression several fetal heart markers in CM clones #19 and #27. The results are expressed as fold change relative to H9 CMs. (E) Immunostaining of cTnT showing significantly enlarged size of GLA-null CMs compared to H9 CMs. (F) Quantification of area size of GLA-null CMs (green columns) and H9 CMs (blue columns). At least 200 cells were analyzed individually and statistical difference is p < 0.001.