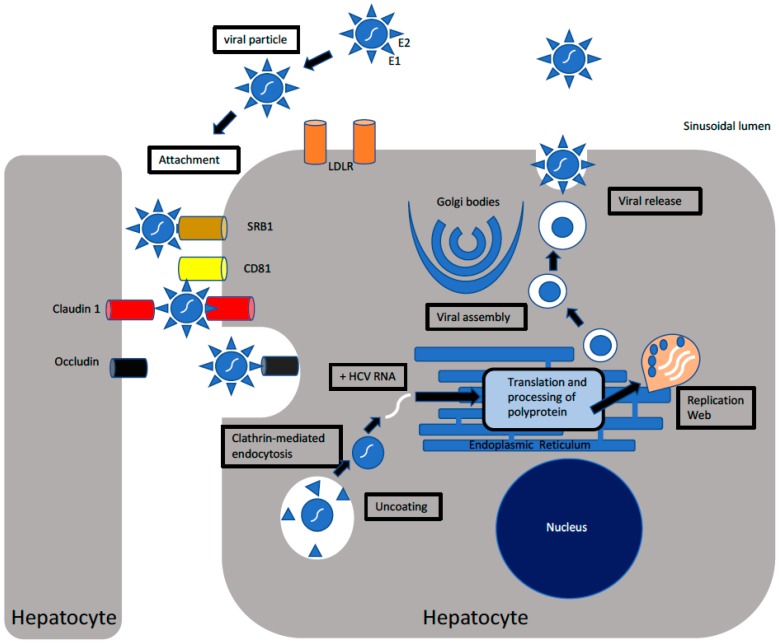
Figure 1.
The replication of hepatitis C (HCV): The virus via its envelope glycoproteins attach to host cellular receptors such as claudin-1, epidermal growth factor receptor (EGFR), scavenger receptor class B type 1 (SRB1), cluster of differentiation (CD81), low density lipoprotein receptor (LDLR), and DC-SIGN (Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin) to attach and subsequently gain entry into host cells. Following attachment, HCV entry occurs via clathrin-mediated endocytosis, wherein HCV undergoes uncoating to release the nucleocapsid into the cytoplasm. HCV RNA is released into the cytoplasm, where it is exposed to host immune machinery. HCV RNA translation via an Internal Ribosome Binding Site (IRES) at the rough endoplasmic reticulum (ER) gives rise to a large polyprotein that undergoes processing into nonstructural and structural proteins. Nonstructural protein NS4B induces the formation of a membranous replication web, where viral RNA replication occurs via the action of RNA-dependent RNA polymerase. The nascent positive sense RNA genome is used for the production of viral proteins, further RNA replication, or the formation of new virions. Utilization of fatty acid pathways along with structural proteins culminate in viral assembly and release.