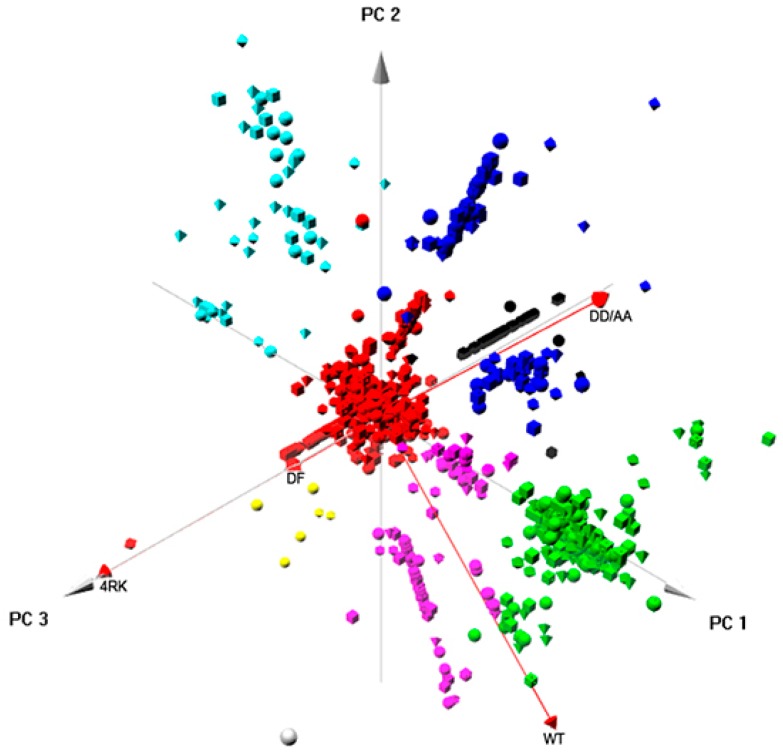
Figure 6.
Component analysis for full-length CFTR variants. PCA was performed using the package “pca3d” from R programming [31]. Data corresponding to the combinations of the median of the amount of each protein immunoprecipitated with each CFTR variant calculated as described above were used for PCA analysis. The PCA scores corresponding to the three main components are plotted in a scatter plot in which the position along the axes show the PCA score of the protein. Clustering was performed using hierarchical agglomerative clustering through the standard R function “hclust” and using average linkage as a clustering method.