Figure 1.
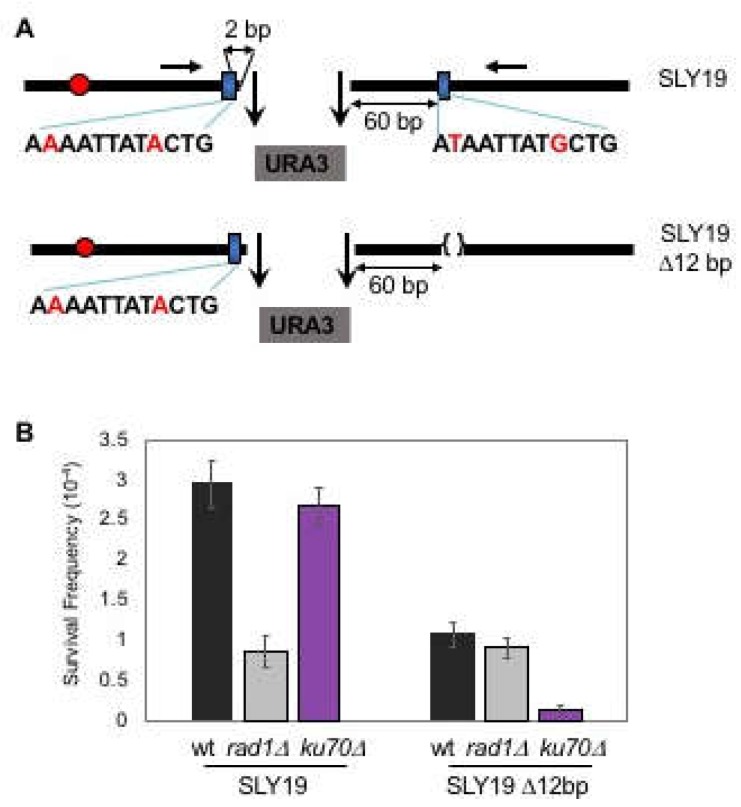
Microhomology selection for microhomology-mediated end joining (MMEJ) is non-random. (A) Two homothallic switching endonuclease (HO) cleavages at the MAT locus (the 117-bp MATa and the full-length MATα cut sites) separated by ∼2 kb of URA3 sequence preferentially induced Ku and RAD52-independent Microhomology-mediated end joining (MMEJ) using the flanking 12-bp of imperfect microhomology that was 2-bp and 60-bp away from the break. The location of the centromere (red circles) and the preferred microhomology (blue boxes) for MMEJ are shown. The sequence (mismatches are shown in red characters) of the preferred microhomology are shown below the blue boxes. The positions of the primers for PCR amplification and sequencing are shown by red arrows. (B) Survival frequency after induction of HO breaks of the wild type (WT), the RAD1 gene deleted [RAD1Δ representing the frequency of classical non-homologous end joining (C-NHEJ)], and the KU70 deletion derivatives (ku70Δ, the frequency of MMEJ) of SLY19 or SLY19 lacking preferred 12-bp imperfect microhomology (SLY19 Δ12-bp). The frequency of survival after an HO-induced double strand breaks (DSB) was calculated by dividing the number of colonies growing on YEP-agar medium containing galactose (YEP–GAL) by the number of colonies growing on YEP-agar medium containing glucose (YEPD). Each value represents the average from at least three independent experiments ± standard deviation.