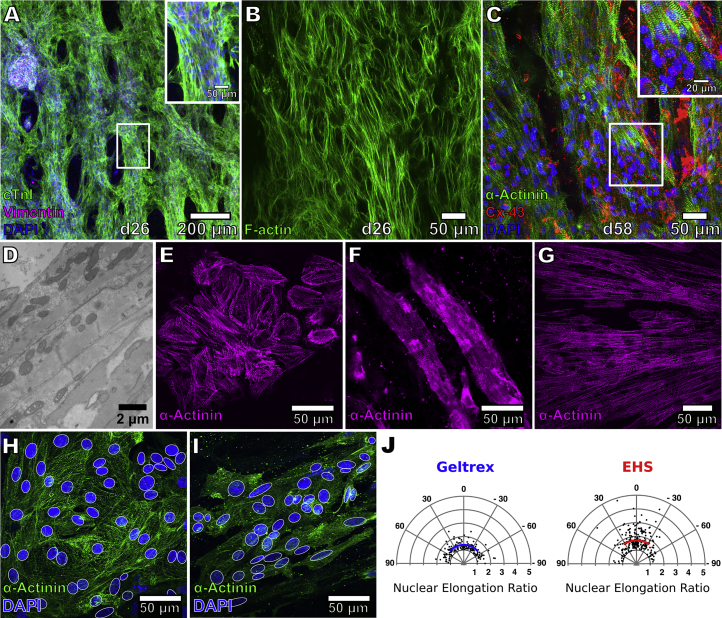
Figure 2.
Morphology of hiPSC-CMs on EHS
(A) Immunostaining for cardiac troponin I (green), DAPI (blue), and vimentin (magenta) in EHS. Scale bar, 50 μm (inset).
(B) Staining for F-actin in EHS.
(C) Staining for α-actinin (green) and Cx43 (red) in EHS. Scale bar, 20 μm (inset). Higher-magnification view in the insets correspond to region outlined by white rectangle. hiPSC-CMs were cultured in EHS for either 16 or 48 days (d26 or d58 of differentiation, respectively).
(D) Transmission electron micrograph of hiPSC-CMs in EHS showing myofilaments and mitochondria.
(E and F) Staining for α-actinin in hiPSC-CMs cultured at a low density on Geltrex (E) or on decellularized slices (F).
(G) Staining for α-actinin in hiPSC-CMs seeded on slices at d13, dissociated after prolonged culture (139 days) and replated on Geltrex.
(H and I) DAPI staining of nuclei (blue, delineated by white outline) in hiPSC-CMs (green, α-actinin immunostaining) on Geltrex (H) and on EHS (I).
(J) Analysis of d24 hiPSC-CM nuclear shape and orientation in EHS and on Geltrex. Radial distances of data points reflect elongation ratios (long axis/short axis) of nuclei that were fitted by white ellipses in (H) and (I). Solid blue and red lines indicate mean nuclear elongation ratios for cells on Geltrex (n = 165 nuclei) or in EHS (n = 233 nuclei), respectively, while arc lengths correspond to standard deviation from the mean angle of orientation.
See also Figures S2 and S3.