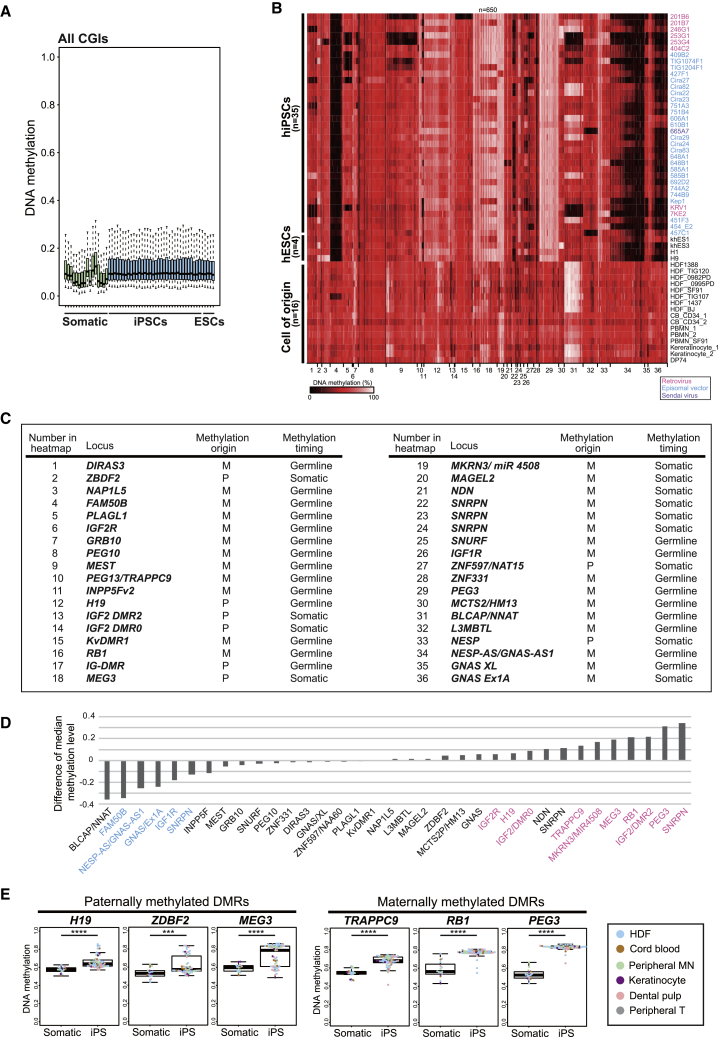
Figure 4.
Variable ICR Methylation Aberrations in Human PSCs
(A) Box plot of DNA methylation levels at all CGIs in human somatic cells and PSCs. Solid lines in each box indicate the median. The bottom and top of the boxes are lower and upper quartiles, respectively. Whiskers extend to ±1.5 IQR. Infinium 450K data of human somatic cells and PSCs were obtained from GEO: GSE60821 and GSE60923.
(B) Heatmap for DNA methylation levels at imprinted DMRs in human iPSCs (hiPSCs), human ESCs (hESCs), and various somatic cells of origin for hiPSCs. Color scale is shown for DNA methylation levels. Infinium 450K data of 35 hiPSC lines (20 male lines and 15 female lines), 4 hESCs, and 16 somatic cells were obtained from GEO: GSE60821 and GSE60923. Names of the hPSC lines are shown at the right of the panel. Colors depict the methods of reprogramming. HDF, human dermal fibroblasts; CB, cord blood cells; PBMN, peripheral blood mononuclear cells; DP, dental pulp cells.
(C) List of 36 human imprinted DMRs shown in (A) (Court et al., 2014). Origin (maternal or paternal allele) and timing (germline or somatic) of methylation are shown for each DMR. IG-DMR (17) is not considered in further analyses because the probes of Infinium 450K are not designed at IG-DMR.
(D) Difference of median methylation levels at the 35 imprinted DMRs between hiPSCs and somatic cells. The median methylation levels of the 35 iPSC lines and 16 somatic cells in (B) are compared. Hypermethylated DMRs (false discovery rate [FDR] <0.01, median methylation in iPSCs >60%) and hypomethylated DMRs (FDR <0.01, median methylation in iPSCs <40%) are shown in pink and blue, respectively.
(E) Box plots of DNA methylation levels at representative paternal and maternal imprinted DMRs in human iPSCs. Solid lines in each box indicate the median. The bottom and top of the boxes are lower and upper quartiles, respectively. Whiskers extend to ±1.5 IQR. Each color in the boxes represents a cell of origin. ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 (Mann-Whitney U test).