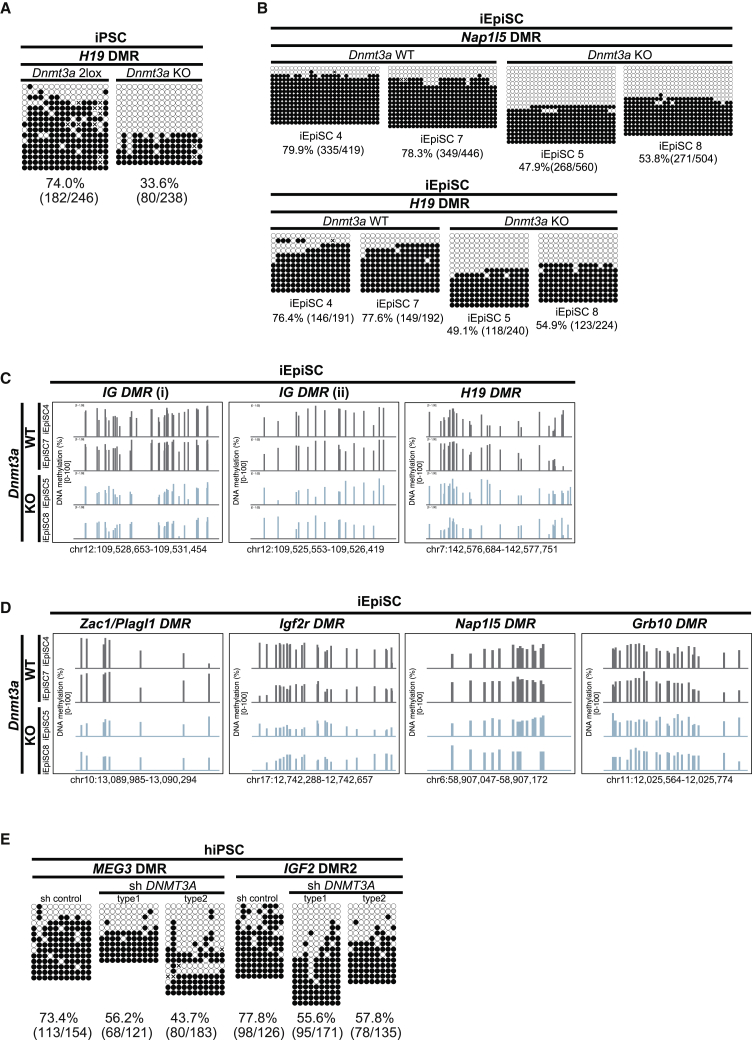
Figure 5.
Dnmt3a Mediates Reprogramming-Associated De Novo ICR Methylation in Mice and Humans
(A) DNA methylation status at H19 DMR by conventional bisulfite sequencing in Dnmt3a control (2lox) and KO iPSCs.
(B) DNA methylation status at Nap1l5 DMR and H19 DMR by conventional bisulfite sequencing in Dnmt3a wild-type (WT) and null (KO) iEpiSCs.
(C) DNA methylation status at paternally methylated DMRs in Dnmt3a wild-type (WT) and null (KO) iEpiSCs. Each bar indicates a CpG site, and bar height represents methylation percentage (0%–100%) by MethylC-seq.
(D) DNA methylation status at maternally methylated DMRs in Dnmt3a wild-type (WT) and null (KO) iEpiSCs. Each bar indicates a CpG site, and bar height represents methylation percentage (0%–100%) by MethylC-seq.
(E) DNA methylation status at MEG3 DMR and IGF2 DMR2 by conventional bisulfite sequencing in hiPSCs established with DNMT3A short hairpin RNA (shRNA) and control shRNA treatment.