Figure 1.
Temporal Expression of OLIG2 in hPSC-Derived VFOs and DFOs
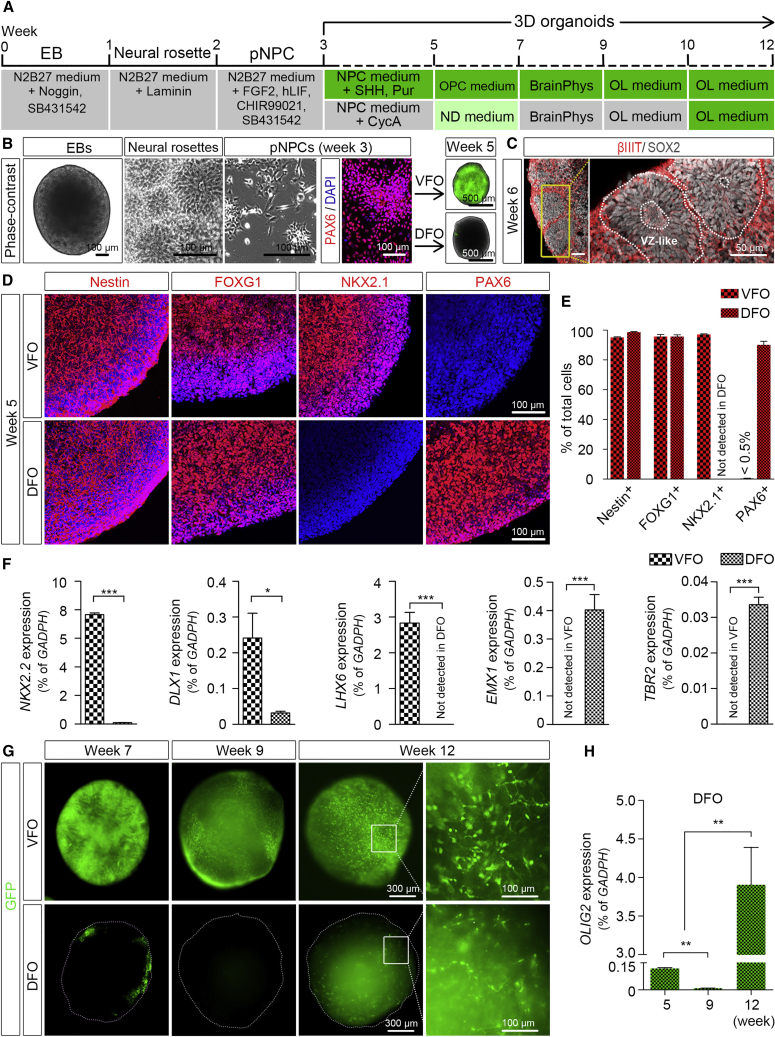
(A) A schematic procedure for deriving brain region-specific forebrain organoids from OLIG2-GFP hPSCs by the treatment of a combination of sonic hedgehog (SHH) and purmorphamine (Pur) or cyclopamine (CycA) alone for VFOs and DFOs, respectively. The stages after week 3 are color coded based on the expression of GFP.
(B) Representative bright-field and fluorescence images of embryoid bodies (EBs) at week 1, neural rosettes at week 2, primitive neural progenitor cells (pNPCs) at week 3, and VFOs and DFOs at week 5. pNPCs at week 3 were positive for PAX6 staining. Scale bars, 100 μm for bright-field images and 500 μm for fluorescence images.
(C) Representatives of the ventricular zone (VZ)-like structure formed by βIIIT+ and SOX2+ cells in DFOs at week 6. Scale bars, 50 μm.
(D and E) Representatives (D) and quantification (E) of Nestin-, FOXG1-, NKX2.1-, and PAX6-expressing cells in week-5 VFOs or DFOs (n = 4 organoids from two hPSC lines).
(F) qRT-PCR results showing the expression of NKX2.2, DLX1, LHX6, EMX1, and TBR2 in week-5 VFOs and DFOs (n = 3 independent experiments). Student's t test: ∗∗p < 0.05 and ∗∗∗p < 0.001.
(G) Temporal expression of GFP fluorescence in VFOs and DFOs. Scale bars, 300 μm in the original images and 100 μm in the enlarged images.
(H) qRT-PCR results showing the expression of OLIG2 at different time points in the DFOs. The expression level is normalized to GAPDH (n = 4 independent experiments). One-way ANOVA with Turkey's post hoc test: ∗∗p < 0.01.