Introduction
Vibrational spectroscopy and imaging promise molecular information that can be rapidly acquired without the need for specialized stains or dyes, thereby potentially simplifying and speeding up necessary analyses for interventions in many facets of modern day healthcare. The salient characteristics of vibrational spectroscopy for molecular analyses, using non-perturbative optical measurements, and employing computational analysis of data, are especially useful near the point of care as assessments can be made with fewer reagents, under pressure of time and accuracy while not requiring extensive specialized human expertise. Significant technological development has occurred and many seminal proof of concept studies have been conducted to demonstrate the utility and vast potential of spectroscopic methods. Accordingly, a number of studies have focused on pushing the fundamental performance limits of spectroscopic methods while others have focused on specific problems where the use of vibrational spectroscopy promises to change the standard of care. Despite this impressive progress, however, the application area is still maturing and rapidly evolving. A vast array of potential applications continues to be assessed while others need further technological developments. In this review, we focus on recent developments that demonstrate potential for point of care impact and major trends that can lead, in turn, to improved spectroscopic technology. We provide focused examples of ‘‘case studies’’ and major trends in spectroscopic analyses ranging from in vivo measurements to that of ex vivo bodily fluids to extracted and processed tissues. In each case, the uniting theme is that information to the clinician is enabled closer to the patient, allowing for a shorter time between identification of the need for analyses and availability of information that guides care.
Raman Spectroscopy for Intraoperative and In Vivo Diagnostics
The vast majority of current methods of assessment are slow, requiring preparation, labeling and human read out. Measurements of intact tissue in vivo promise the least disruption to the person being diagnosed, which eliminates the many steps needed in assessments. One of the major advantages of vibrational spectroscopy techniques for in vivo measurements, in addition to the molecular measurement of disease,1 is the immediacy of the diagnostic information that can be obtained with little or no sample preparation. This means that molecular diagnostic approaches without recourse to labels or stains can be available to the clinician at the point of care. In vivo measurements can also help guide when a sample of bodily fluid, a cytology (cellular) sample or a biopsy (tissue) sample are taken, either for a definitive diagnosis or to triage a patient into a subgroup of at risk patients for further analysis. Sampling is the current standard of care and the acquisition, handling and analyses processes of current methods can lead to a high level of inter- and intra-observer disagreement.2 Vibrational spectroscopy can provide an objective and rapid measure of the molecular composition and, coupled with multivariate analysis or machine learning approaches, can provide an accurate prediction of disease state. While this opportunity is immense, several challenges also need to be overcome to realize the potential of in vivo measurements.
In Vivo Raman Probes
The major technological challenges of in vivo use of vibrational spectroscopies lie in the limited penetration depth of mid-infrared (mid-IR) light in water rich tissues and the relatively weak signals from Raman scattering, which can be compromised by background signals from measurement systems, scattering, and/or fluorescence. Raman probes can be utilized as clinical tools able to provide rapid, noninvasive, real-time molecular analysis of disease specific changes in tissues. Clearly the target tissue location, the significance of spectral changes with disease and the possible access routes to the region of interest will vary for each clinical application considered. Most Raman applications seek to measure the fingerprint region of the spectrum ~400–1800 cm‒1 where the greatest abundance of inelastic scattering peaks from biomolecules are found. There are several challenges and proposed solutions to recording the data. Use of near-infrared laser illumination can minimize tissue fluorescence contributions in this region, but signals from the silica fibers can dominate fingerprint Raman spectra and need careful optical design and filtration strategies. For example, these signals can be subtracted but it is not possible to subtract the accompanying shot noise. Since the Raman effect is weak, noise may be sufficiently large to impact the relatively small Raman signals obtained in the ideally short time scales required for in vivo measurement. Silica signals in the illumination fibers are usually filtered prior to the light reaching the sample, using a band pass filter and elastically scattered light from the sample filtered at the probe tip using a long pass edge filter or notch filter, allowing only the Raman signal to pass along the collection fibers to the spectrometer, thus minimizing any further signal contamination.
An alternative method has been explored for filtering the elastically scattered laser light from the collection fibers and thus minimizing the induction of background signal from fibers. This uses in-line fiber Bragg gratings (FBG) to reject/reflect the laser light in the collection path. A Raman probe was built consisting of one excitation fiber and six multicore single-mode fibers (19 cores) with inscribed FBGs as collection fibers.3 A more simple approach to overcome this problem is to use only the high wavenumber region of the spectrum, reducing the impact of the background from the fibers and allowing for simpler and cheaper unfiltered probes.4 However, the limited spectral features in this region 2400–4000 cm−1 may limit the diagnostic capability of the system to extreme pathologies such as invasive cancer versus normal healthy tissues. For instrumentation, there is always a trade-off between the magnitude of signal recorded, the time required and the cost of hardware to do the same. This range presents opportunities for spectroscopists to devise solutions that provide measurements in an efficient and accurate manner.
In addition to design and trade-off considerations of instrumentation, the samples provide another class of challenges. One of the most significant requirements of in vivo Raman systems relates to the specific sampling volume of the clinical application and sampling method. Near-infrared (NIR) light is highly scattered and minimally absorbed in tissue, and therefore optical designs that do not restrict light collection to the surface scattered photons can result in deep signal collection, which may confound results. An example of this is when no beam steering or focusing is used in hollow organ endoscopic fiber probes. The relevant diagnostic signal for dysplastic (early cancerous) lesions may originate from the surface 100–200 μm in organs such as the esophagus or bladder, and any deeper signals may contain contributions from, for example, normal cell division and adipose tissue on the outer organ surface.5 It is not a trivial process to build a complex Raman probe for use down a working channel of an endoscope, particularly when one considers the total diameter should be less than 2–3 mm and these contain lenses and filters requiring careful alignment and packaging. A recent review provides more detail on the strategies to optimize in vivo Raman measurements with Raman probes.6
An important consideration for in vivo measurements is the need to minimize instrumentation size such that the subjects being measured are inconvenienced the least and smaller portions of the body can also be addressed. Miniaturization considerations lead to the prospect of Raman needle probes enabling rapid analysis of disease specific molecular changes in deeper tissues such as lymph nodes, breast and prostate. A smart Raman needle probe has been developed and tested for potential in vivo and ex vivo use, capable of measuring tissue Raman molecular tissue signals and demonstrating spectral differences between metastatic and non-metastatic nodes in <1–2 seconds down a hypodermic needle.7 Further developments have led to a hand-held device containing the key optical components coupled to disposable needle probe tips.8 Initial ex vivo feasibility testing of the technique was performed on excised head and neck lymph nodes from 62 patients undergoing surgery, covering all pathologies observed in this sequential case mix, including reactive, lymphomas and secondary cancers. Initial results show reasonable performance with an area under the receiver operator curve of 0.83 for malignant versus non-malignant nodes.
Deep Raman Approaches
Beyond accessing disease specific molecular signals using fiber probes, the rapidly developing technique of deep Raman spectroscopy is emerging as a powerful in vivo tool. Early studies, in pioneering the field of deep Raman spectroscopy for biomedical applications, have established the basic feasibility of recovering Raman signals (both native and exogenous) from depths of several centimeters beneath the surface of animal tissues. This is, by around two orders of magnitude, deeper than achievable with conventional approaches such as confocal Raman microscopy. Deep Raman sampling involves the use of either transmission (TRS) illumination and collection geometries, or spatially offset Raman spectroscopy (SORS), whereby illumination and collection points are spatially separated on the outer surface of the tissue.9–11 Initial work has involved the study of signals from substances such as those found in calcified tissues, whereby the signal is distinct from that found in soft tissues. Clinically relevant concentrations of calcifications have been detected to depths of up to 40 mm in animal tissue phantoms (i.e., at near clinically relevant depths; X-ray mammographic screening compression ranges from 1.9 to 5 cm thick) demonstrating the basic viability of scanning human breasts.12 This penetration depth was accomplished by dramatically increasing the Raman photon gathering rates (by around 100 times) when compared with the first TRS Raman system used.13 This indicates the feasibility of using TRS for noninvasive analysis of breast tissues in vivo.
In Vivo Application Examples
Skin.
The most common cancers are found in the skin, the largest and most accessible organ in the body. Numerous studies have explored the Raman analysis of skin in vivo. Lieber et al used a Raman fiber probe to measure lesions from non-melanoma skin cancers and demonstrated 100% sensitivity and 91% specificity in discriminating these lesions from normal tissues in this small study.14 A much larger study (over 1000 cases), using a hand held Raman probe measuring spectra in less than 1 s, was able to identify malignant melanoma lesions with high sensitivity >90% but low specificity, ranging between 15% and 54%, when discriminating malignant melanoma from non-melanoma pigmented lesions and seborrheic keratosis.15 A combination of both Raman and optical coherence tomographic (OCT) imaging has been explored to provide both tissue architectural features and biochemical signatures from the same locations in skin. The system uses independent light sources 785 nm and 1300 nm detection systems for Raman and OCT respectively and shows some promise for distinguishing basal cell carcinomas from surrounding normal skin.
Bone.
Pioneering work by Matousek and Morris in developing SORS16 has enabled the possibility of measuring bone composition in vivo.17 These developments have mostly been focused on the perspective analysis of osteoporosis, although other conditions are also being explored. Esmonde-White et al., showed that when measuring bone composition with Raman spectroscopy, that dicalcium phosphate dihydrate and uncarbonated apatite were found to be associated with infected bone as opposed to carbonated calcium hydroxyapatite in healthy bone.18 This indicated that an in vivo measure of bone composition could provide a measure of infection. Figure 1 shows an in vivo SORS measurement using a handheld SORS probe (built in house, Stone lab) from the knuckle in 1 s.
Figure 1.
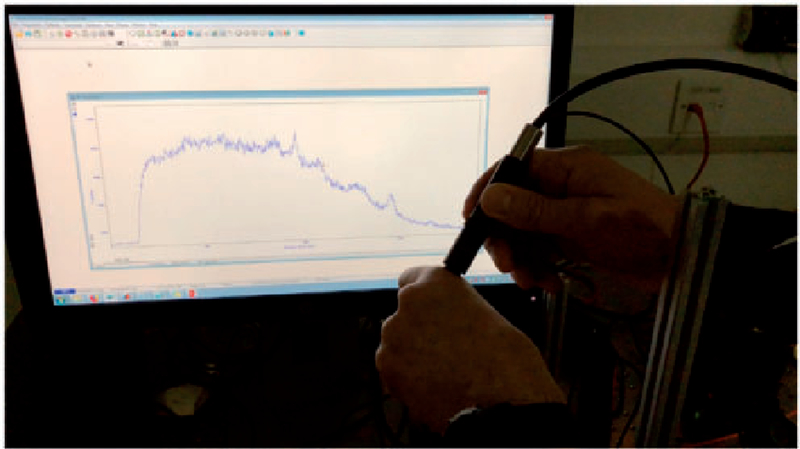
Measurement of SORS spectrum of the knuckle using 830 nm illumination and a hand held SORS probe. Peaks show hydroxyapatite of bone, and protein peaks. Collection time 1 s per spectrum. Unpublished data from N. Stone labs.
Endoscopic Disease Detection in Hollow Organs
The gastrointestinal tract includes organs (esophagus, stomach, colon) that develop many of the most significant cancers with high incidence, mortality and often a slow genesis. This allows for the possibility of early detection and effective treatment. However, current methods of sampling and identifying microscopic lesions are limited.2 Early work by Shim et al.19 showed some potential, but insufficient signal to noise was obtained for early diagnosis of malignancies. The team of Stone et al., have been devoted to developing and testing a confocal probe able to measure signals from only the surface 100–200 μm of the esophageal mucosa,20,21 shown to be the optimum depth for identification of early dysplastic changes leading to cancers.22 Seven hundred and ninety-eight, one second Raman probe spectra were acquired from 673 esophageal tissue samples from 62 patients. Acquisition times between 5 s and 0.1 s per spectrum were evaluated. Principal component fed linear discriminant analysis was used to calculate probe accuracy by reference to a consensus histopathological ‘‘gold standard’’ diagnosis. All results were statistically cross-validated based on characteristic spectral signatures. High-grade dysplasia and adenocarcinoma could be discriminated from Barrett’s esophagus, low-grade dysplasia, and normal squamous esophagus with a sensitivity of 86% and a specificity of 88%. The ability to detect early superficial mucosal disease, including discrimination between low-grade and high-grade dysplasia (HGD), was also demonstrated despite short, clinically applicable (1 s) spectral acquisition times. However, enhanced diagnostic accuracy was demonstrated when using 5 s acquisition times; the detection rate of HGD–adenocarcinoma remained 86%, but the specificity was greatly improved at 98%.20
Huang et al. have demonstrated various applications of in vivo Raman diagnostics, particularly in the gastrointestinal tract.23,24 They have been exploring different probe configurations to provide diagnostic signals from the lining of hollow organs such as the oesophagus.25 These multifiber probes have been shown to be efficient in terms of light collection, and ongoing work has enabled more specific sampling of diagnostically relevant surface signals. Following on from early work by Stone et al. demonstrating Raman discrimination of dysplasias and cancers in the larynx,26 Lin et al. have also recently demonstrated in vivo diagnosis of laryngeal carcinomas.27 In lung cancer diagnostics, a bronchoscope based on white light and autofluorescence was integrated with a Raman probe system to explore the value of Raman in improving the specificity of pre-neoplastic lesion detection.28 White light and autofluorescence images allowed identification of suspicious lesions, areas from which Raman spectra were measured with acquisition times of 1 s. The authors showed that sensitivity of 96% and a specificity of 91% for discrimination of pre-neoplastic lesions (leave-one-out cross-validations) by developing multivariate statistical models.
Intraoperative Raman Analysis
Raman measurements have been proposed by a number of teams for intraoperative analysis to provide the surgeon with a real-time measure of tumor margin analysis or metastatic lesions such as sentinel lymph nodes. The field of Raman spectroscopy for cancer detection and cancer surgery guidance has recently been reviewed by Santos et al., demonstrating real progress towards clinical translation of these techniques and adoption by the medical community, as well as highlighting challenges.29 A sequence of studies have shown that Raman spectroscopy can accurately identify metastatic invasion in lymph nodes from the axilla, mediastinum, and head and neck (H&N) using Raman microscopic mapping of the cut surface of the nodes.30–32 NIR Raman has been demonstrated to identify the pathology of swollen lymph nodes from the head and neck. The collected Raman spectra can clearly be separated into those from reactive nodes (swollen from reaction to infection), primary malignancies (lymphomas) and secondary malignancies (metastatic squamous cell carcinomas and adenocarcinomas) with 90% sensitivity and 86% specificity.33
This performance has been reproduced in axillary nodes during breast surgery, using a low-cost commercially available, portable probe-based Raman system to measure the molecular fingerprint of the excised sentinel lymph nodes (SLN) as shown in Figure 2. Greater that 85% sensitivity and 96% specificity was achieved for identification of metastatic nodes. This study was limited by the sample volume measured (using the off the shelf probe), which led to a small number of micrometastases (lesions smaller than 1 mm) being missed. Customized sampling of larger volumes is expected to improve this performance further. The performance of non-optimized Raman in the breast SLN feasibility study versus other intra-operative results (Table I) showed Raman measurements to be equivalent to the molecular assay approaches, but they require no tissue destruction (allowing for follow up histopathology). Furthermore, Raman provided more rapid results and could be applied in vivo.34
Figure 2.

Sentinel lymph node identified using radiotracer and blue dye during surgery, the excised node is placed on the end of a Raman handheld probe and the spectra measured show clear differences between infiltrated and non-infiltrated (metastatic) nodes.
Table I.
Performance of range of intraoperative techniques versus nondestructive Raman measurements near patient.
Margin Analysis
Raman spectroscopy has also been proposed for detection of tumor margins during breast cancer surgery, both in vivo and ex vivo. A handheld Raman probe was developed for in vivo collection of single-point Raman spectra during surgery.42 This was tested in nine patients undergoing partial mastectomy procedures providing tissue spectra at each measurement point in 1 s. Spectral data was fit with basis spectra from tissue components to provide biochemical profiling.43 From a clinical perspective this approach is unlikely to yield sufficiently rapid sampling of all margin locations in vivo. Extensions of this work utilizing the relatively new approach of SORS to sample up to 2 mm beneath the resected sample surface has enabled an intraoperative approach measuring margins on ex vivo tissue to be explored.44
Another potentially important intraoperative application is Mohs surgery, whereby basal cell skin cancers are excised in a slice by slice manner until no tumor is found on histopathological assessment. This is a particularly time consuming and costly process. The use of wide field autofluorescence imaging of skin tissues to enable rapid identification of regions of concern for localized Raman measurements during Mohs surgery shows great promise for enabling dermatological surgeons to obtain an accurate measure of basal cell carcinoma margins in the operating theatre, rather than waiting for pathological analysis.45 The approach could provide a particularly valuable adjunct to current techniques. In the field of oral cancer, Barroso et al.46 have demonstrated discrimination between oral cancer and healthy tissue based on water content of freshly excised tongue specimens, determined by Raman spectroscopy in the high wavenumber region. This dramatic signature difference can be used to determine the location of the tumor border in oral cancer surgery.47
There has been a recent burst of activity in the exploration of Raman as a tool to support brain conserving cancer surgery. Identification of tumor margins is critical to minimize the potential for recurrence, and this is particularly difficult for gliomas, which have poorly defined margins. The benefit of minimizing the amount of tissue removed is obvious, particularly in the case of brain surgery, and therefore the additional guidance of a spectroscopic probe to detect invasive brain cancer in situ in real time in patients is potentially invaluable. Jerym et al. demonstrated the use of a handheld contact fiber optic Raman spectroscopy probe to distinguish brain cancer (glioma) from normal brain in situ with sensitivities and specificities exceeding 90%.48 In a study of 35 patients, Vaqas et al. similarly deployed Raman spectroscopy to analyze tissue at the brain surface, en route to the tumor, in superficial and deep tumor zones and resection margins in vivo during surgery, demonstrating that, in comparison to matched core biopsy samples verified by routine histopathology, meningiomas, and metastases, as well as low- and high-grade gliomas could be identified with high accuracy.49 These and other recent developments of applications of Raman based spectroscopy for improving the accuracy of brain tumor surgery have been reviewed by Hollon et al.50
Summary and Outlook for In Vivo Measurements
Although in vivo Raman shows great promise and many positive results, nearly all studies to date are statistically under powered. This is often due to the constraints of running studies at single centers with limited patients numbers and funding. A step change is needed to ensure sufficiently large numbers of patients can be recruited to build diagnostic models able to describe the majority of the variance in the population of interest as well as to test them. Furthermore, it is vital that more than one pathologist, ideally at least three is used to provide a better than gold standard ‘‘ground truth’’ to ensure that inter- and intra-observer differences in pathology do not confound the results. Note also the additional value of molecular spectroscopic to go beyond replicating a measure of the gold standard or immunohistochemical markers. It has a huge potential to provide a genuine measure of likely prognosis for the patient from the first measurements. Crow et al. showed bladder cancer surface measurements were indicative of the stage as well as grade of disease, i.e., the level of invasion into the surrounding organ as well as the cancerous nature if the individuals cells being probed.51 Kendall et al., showed differences in high grade dysplasia signals at first presentation between those progressing early and late to invasive adenocarcinoma.52 Leiber et al., showed in organotypic skin tissue raft cultures that Raman could distinguish the presence of sarcoma fibroblast cells in regions adjacent to the sampled volume, when compared to normal fibroblast cells.53 Furthermore, Singh et al. demonstrated malignancy associated changes in the buccal mucosa probed by Raman in at risk patient groups from tobacco exposure.54 In summary, Raman spectral measurements show a high potential for specific applications and new areas of application can emerge with the practical measurement technologies. Together, these applications not only provide a benefit in providing currently known diagnostic information but can also provide new means of understanding disease progression and risk in the future.
Raman Spectroscopy and Imaging for Ex Vivo Samples
Raman spectroscopy is a highly valuable tool regarding the analysis of biological samples, providing detailed insight into their chemical composition. In addition, due to its nondestructive nature and speed, Raman spectroscopy has been recognized as promising technique in medical diagnostics for many years, as detailed for in vivo measurements in the previous section. Over the last years, increased efforts have also been made towards establishing Raman spectroscopy also in point-of-care diagnostics, where the demands in terms of robustness, user friendly handling and cost efficiency are even higher. A particularly relevant area in point-of-care applications is the diagnosis of infectious diseases. Not only will the patients directly benefit from a rapid diagnosis, because an appropriate medication can be administered in a timely manner, but also a responsible management of antibiotics is enabled. Since the occurrence of antibiotic resistances increases at an alarming rate and the concern of approaching a post-antibiotic era respectively grows, the strict practice of antibiotic stewardship is long overdue. However, the corresponding measures can only be efficiently realized, if fast and reliable tests are widely available for determining the cause of infection and possible resistances.
To illustrate the potential for point of care applications, this section will focus on recent developments in applying Raman spectroscopy for diagnostics of infectious diseases. As depicted in Figure 3, Raman spectroscopy offers various options for addressing this task. Special attention will be paid to the investigation of easily obtainable body fluids, such as urine, blood, saliva or sputum, as they are the most suitable sample types in point-of-care assays in contrast to cerebrospinal fluid55–57 or bronchoalveolar lavage, which require invasive procedures. Analyzing highly complex samples like body fluids using Raman spectroscopy entails certain challenges: depending on the assay more or less complex sample preparation strategies need to be applied and statistical analysis of the acquired data often is inevitable. Another core theme is the identification of antibiotic resistances, since this information is vital for the treating physician and highly significant for containing the unnecessary and problematic use of antimicrobial drugs.
Figure 3.
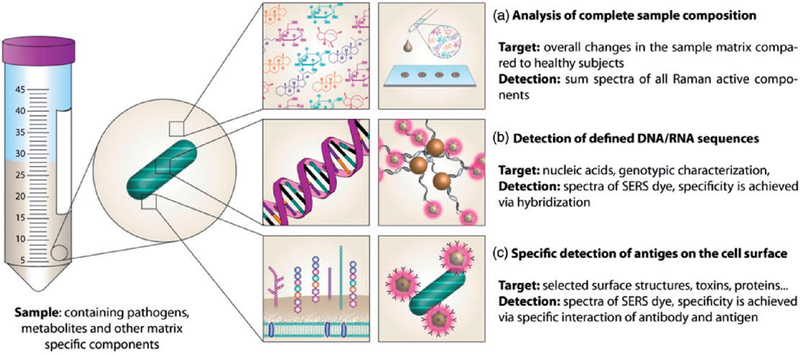
Schematic display of different approaches for detecting infectious diseases using Raman spectroscopy.
Detection of Pathogens in Urine Samples
Urinary tract infections (UTI) are a very common disease, especially among women. Often their progression is harmless and easily treatable. However, serious complications can arise, if no appropriate medication is given or if the immune system of the patient is otherwise compromised.58 Bacterial cell concentrations of urine samples from patients with UTI can range from 102 to 105 cells/ml.59 Due to the complex chemical composition of urine60 a sample preparation strategy before the Raman spectroscopic investigation of the pathogens is generally required in order to prevent the Raman active compounds of the urine matrix to interfere with the bacterial spectra. Most commonly, centrifugation is employed in order to separate the bacterial cells from the sample matrix. Furthermore, washing steps with buffers follow, so that residual contaminants can be removed from the cells. Sometimes a filtration step is included for eliminating possible eukaryotic cells such as leukocytes or epithelial cells prior to the centrifugation procedure. The isolated bacteria can be either investigated as bulk sample or as single cells. As illustrated in Figure 4, each method has unique advantages that can help in different analytical situations, from measurements of single cells to colonies, as well as trade-offs in terms of preparation of samples and time until which results may be available.
Figure 4.
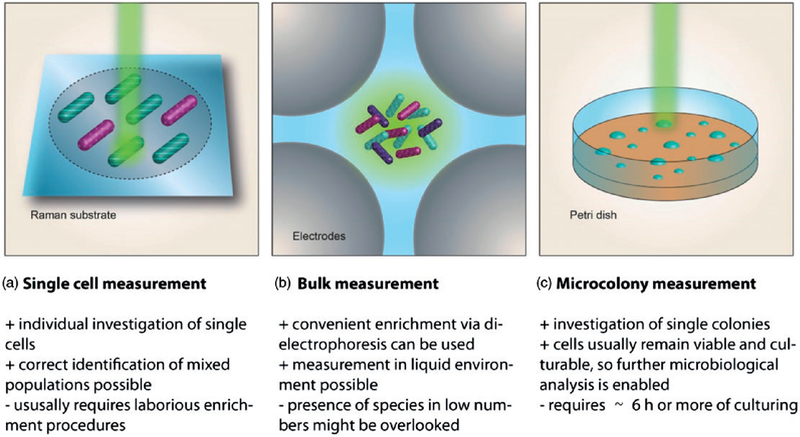
Schematic display of different modes for investigating bacterial cells using Raman spectroscopy.
Several exciting innovations have been made in this area. Schroeder et al. introduced a lab-on-a-disc platform, which enables a convenient enrichment of bacterial cells from urine samples.61 After a short pre-treatment step, involving filtration and volume reduction, the sample is transferred into the microfluidic disc, which is then placed on a rotator. Due to the centrifugal forces, the bacterial cells are collected in micrometer-sized wells. The Raman spectroscopic investigation of the bacterial cells in bulk mode can be conducted directly in the disc platform (Figure 4b). The applicability of the system was demonstrated by analyzing samples of patients with significant bacteriuria (105 CFU/ml or higher) caused by Escherichia coli or Enterococcus faecalis. The complete procedure from sample preparation to result required about 70 min, which is a great improvement compared to culture based identification. Neugebauer and coworkers developed a dielectrophoresis (DEP) chip that allows concentrating bacterial cells in liquid samples.62 Subsequently, Raman spectra of the cell cloud can be recorded. The DEP chip was successfully tested with cultured bacteria as well as with actual patients’ urine samples with confirmed UTI. An unambiguous identification of E. coli and E. faecalis was achieved within only 35 min. In contrast to the previously mentioned studies Kloss et al. developed a centrifugation based sample preparation routine optimized for Raman microspectroscopic investigations of single cells (Figure 4a).63 In order to directly identify the bacteria responsible for the infection a database with single cell spectra of eleven different pathogens, relevant for UTIs, was established. Several patient samples were accordingly analyzed, among them also samples from patients that had received a course of antibiotics. All samples were correctly identified within approximately 2 h. The proposed method allows analyzing samples with a concentration of 103 CFU/ml and higher.
Premasiri et al. employed surface enhanced Raman spectroscopy (SERS) for investigating UTI relevant bacteria spiked into urine samples after several centrifugation and washing steps.64 The SERS substrate with Au nanoparticles, providing a strong enhancement of the Raman signal from the bacterial cells, enabled them to incorporate a portable Raman microscope in their system. Catala et al. also chose SERS as detection method for identifying and quantifying Staphylococcus aureus in urine samples, blood and other body fluids.65 They modified Ag nanoparticles with Raman reporter molecules as well as with specific capture probes (antibodies and aptamers) for S. aureus. The biorecognition elements cause the SERS encoded particles to accumulate on the surface of the S. aureus cells, resulting in a strong increase of the SERS signal of the Raman reporter. For quantification, a microfluidic device was developed in which the SERS signal is continuously monitored. This approach allowed detecting cell concentrations of 15 CFU/ml and lower within 20 min.
Detection of Pathogens in Blood Samples
In terms of complexity, blood is an even more challenging sample matrix. The main components are blood plasma and the formed elements, which comprise erythrocytes, leukocytes and platelets. The most abundant molecules, found in the plasma, are various proteins, but enzymes, hormones, amino acids, nutrients as well waste products are present as well. Furthermore, it has to be considered, that gases like oxygen, carbon dioxide and nitrogen are dissolved in the plasma. Detecting pathogens directly in the whole blood matrix, which contains billions of blood cells, is extremely difficult, since quite often there are only 10 CFU/ml or fewer bacteria present. Nevertheless, there is an urgent need for methods that allow rapidly diagnosing life threatening conditions such as sepsis, because nowadays the standard approach is still relying on time consuming blood cultures. In the last years, major efforts have been made to establish sample preparation strategies for blood, which enable using Raman spectroscopy as a diagnostic tool for sepsis or bacteremia.
Boardman et al. developed a preprocessing routine for whole blood that concentrates viable microorganisms from a 10 ml sample to a 200 µl volume.66 They performed a selective lysis step for the blood cells and used a centrifugal concentration device for enrichment. They were able to recover various bacterial species as well as C. albicans from whole blood samples with excellent yields of 55 % and higher. The content of pathogens in the blood samples was either 2 or 10 CFU/ml, which represents a relevant concentration range for sepsis or bacteremia. Prior to the SERS measurements, an incubation step is performed to obtain enough cells for the spectroscopic investigation. The authors successfully demonstrated, by investigating several spiked samples, that their combination of sample preparation and SERS based analysis allows a sensitive and specific identification of bacteria within 7 h. The introduced sample preparation method might be highly useful for other spectroscopic and non-spectroscopic detection methods.
Ngo et al. also developed a SERS assay for analyzing whole blood samples.67 However, they aimed at a nucleic based identification of the malaria parasite Plasmodium falsiparum. Remarkably, they were able to detect the pathogen RNA without using a nucleic acid extraction protocol or amplification step. By employing magnetic beads as platform for a hybridization assay with SERS nanoparticle modified oligonucleotides as reporter probes, they achieved a limit of detection of 200 fM with synthetic target DNA. Further Raman based studies of blood samples concerning the diagnosis of malaria have been reported.68,69 Chen et al. found that SERS spectra of malaria infected red blood cells (RBC) show different features than uninfected cells and also that the stages of infection can be differentiated according to the SERS spectra of the investigated cells. Kozicki et al.69 performed a detailed Raman based analysis of the early ring stage of RBCs from malaria patients. They were able to observe the chemical and structural changes resulting from the parasite infection in the Raman spectra, even though the hemogram did not show any changes in the RBC parameters in the investigated early stage.
Blood serum and plasma are also frequently used in routine clinical diagnostics and can provide valuable information about a patient’s condition. Neugebauer et al. analyzed blood plasma from ICU patients either with systemic inflammatory response syndrome (SIRS) or sepsis using Raman spectroscopy.70 They were able to distinguish between SIRS and sepsis with 80% accuracy, which is very promising, because no distinct sepsis biomarker is known so far. Kamińska developed a SERS immunoassay for detecting interleukin 8 (IL-8), a cytokine associated with inflammation, in blood plasma and improved the limit of detection compared to a conventional enzyme linked immunosorbent assay (ELISA).71 Bonifacio et al. conducted an extensive study with blood plasma and serum and in order to find the optimum experimental conditions for reproducible SERS spectra.72 For example, they found that the anticoagulants ethylenediaminetetraacetic acid (EDTA) and citrate have a much stronger influence on the spectra than heparin. Tatarkovič et al. compiled an experimental procedure for minimizing the fluorescent background in Raman and Raman optical activity (ROA) spectra.73 Furthermore, the capabilities of Raman spectroscopy in diagnosing viral infections from blood serum have been explored. Khan et al. were able to differentiate between dengue virus infected and uninfected samples with 85% diagnostic accuracy and compared the performance of standard ELISAs to the Raman spectroscopic approach.74,75 Anwar at al. were able to detect biochemical changes in blood serum associated with hepatitis B and C.76
Detection of Pathogens in Other Body Fluids
Sputum and saliva belong to the category of easily obtainable samples for point-of-care diagnostics. Kloss et al. established a method for isolating intact single bacterial cells from sputum samples, which involves filtration as well as centrifugation steps, for subsequent Raman spectroscopic investigations. A statistical model for classifying pathogens relevant for lower respiratory tract infection was built. Validation with an independent data set yielded a correct identification rate of 97.4%.77 Gonchukov et al. identified carotenoids as biomarkers for periodontitis and were able to specifically detect them in dried saliva samples from periodontitis patients via resonance Raman spectroscopy.78 Ghebremedhin et al. employed SERS for verifying bacterial presence in wound effluent samples.79
Identifying Antibiotic Resistance
Nowadays, the identification of the bacterial species alone is no longer sufficient and further information about antibiotic resistances is required. This need clearly has been recognized by the Raman spectroscopic community and various approaches have been published. Assmann et al. studied the effects of vancomycin on a sensitive E. faecalis strain using Raman microspectroscopy, while Kang et al. were able to detect carbapenemase activity in the New Delhi metalloproteinase (NDM) producing E. coli strain using SERS.80,81 Schroeder et al. designed a dielectrophoretic chip, which allows Raman spectroscopy monitoring of the phenotypic changes in resistant and sensitive strains of E. coli induced by the antibiotic ciprofloxacin.82 Based on a classification model the identification ciprofloxacin-resistant E. coli is possible within 3.5 h. Kirchhoff et al. demonstrated that Raman spectroscopy is even capable of determining the minimal inhibitory concentration (MIC) of ciprofloxacin.83 By combining dielectrophoretic enrichment and statistical analysis of the bulk spectra, they were able to determine the MIC for various clinical E. coli strains. Dekter at al. conducted an extensive study for examining the effects of various antibiotics on the Raman spectra of eight different sepsis relevant pathogens.84 They further demonstrated that the antibiotic susceptibility detection using Raman spectroscopy also succeeds for bacteria isolated from spiked blood culture samples. Premasiri et al. reported a SERS based method enabling a strain specific differentiation of pathogens directly isolated from urine samples.85 Their study included diverse resistant and sensitive E. coli strains, which could be unambiguously classified according to their specific SERS signature. Novelli-Rousseau et al. investigated the influence of different antibiotics on the Raman signature of susceptible and resistant E. coli strains.86 They were able to identify a signature in the Raman spectra induced by the effect of the antibiotics. Based on the acquired spectra they successfully trained a classifier for detecting antibiotic susceptibility on a single cell level. Within this context, the work of Mathey et al. might be of interest.87 They found that the laser irradiation used in Raman spectroscopic investigations of micro-colonies does cause some membrane damage, however, the colonies still kept their ability to grow and thus remain available for further antimicrobial susceptibility testing (Figure 4c).
Summary and Outlook for Ex Vivo Measurements
Impressive progress has been made towards implementing Raman spectroscopy in medical diagnostics and designing assays suitable for point-of-care applications in recent years. The importance of sample preparation is increasingly appreciated and more studies are published addressing the complete chain of analysis. Even though often preliminary experiments are performed with buffer as sample matrix, researchers frequently spike actual body fluids with cultivated bacteria in order to simulate the patient’s samples as realistically as possible. Table II provides an overview of the different options for sample preparation depending on the sample matrix. Another encouraging development is the fact, that even for highly complex matrices such as blood, efficient sample preparation strategies, which also take the clinical relevant concentration range into account, have been reported. However, there are still some obstacles that need to be overcome before Raman spectroscopy can be employed in routine point-of-care diagnostics. Although well working sample preparation strategies have been realized, in most cases they involve multiple devices and various manual handling steps, so that they only can be performed in a specialized laboratory. For on-site application, an ideally complete automation of the respective protocols is desirable. In principle, this need already has been recognized and efforts have been already made towards this goal. For example, handheld Raman setups are available and have been integrated in the analysis of highly complex samples. In addition, in terms of sample preparation many of the proposed approaches have been automated to some degree and clearly possess great potential for further automation and miniaturization. With these anticipated advances, Raman spectroscopy will become a valuable tool for routine on-site analysis and point-of-care applications in near future.
Table II.
Overview of options for sample preparation strategies for different body fluids enabling Raman spectroscopic identification of infectious diseases.
| Sample type | Pathogens | Sample preparation | Concentration | Time (time for isolation) | Reference |
|---|---|---|---|---|---|
| Urine | E. coli, E. faecalis | Filtration, centrifugation, enrichment via lab-on-a-disc | >105 cells/ml | 70 min (65 min) | 61 |
| E. coli, E. faecalis | Filtration, dielectrophoresis chip | >105 cells/ml | 35 min (15 min) | 62 | |
| P. aeruginosa, K. pneumoniae, E. coli, E. faecalis, E. faecium, S. aureus, P. mirabilis, S. epidermidis, S. haemolyticus, S. hominis, S. saprophyticus | Centrifugation | >103 cells/ml | 2 h | 63 | |
| E. coli, K. pneumoniae, S. saprophyticus | Centrifugation | 105 cells/ml | 30 min | 64 | |
| S. aureus | SERS-encoded Ag nanoparticles, microfluidic platform | >15 CFU/ml | 20 min | 65 | |
| Whole blood | E. coli, K. oxytoca, S. aureus, P. aeruginosa, S. pneumoniae, H. influenza, C. albicans | Selective lysis, centrifugation, cultivation | <10 CFU/ml | 7 h | 66 |
| P. falsiparum | Magnetic bead-based hybridization, lab-on-a-stick platform | 200 fM | not specified | 67 | |
| Sputum | S. aureus, S. epidermidis, S. pneumoniae, K. pneumoniae, P. aeruginosa | Enzyme based liquefaction, filtration, centrifugation | >104CFU/ml | (1 h) | 77 |
Point-of-Care Applications of Infrared Spectroscopy and Imaging
Fourier transform infrared (FT-IR) and discrete frequency IR (DF-IR) imaging using quantum cascade laser (QCL) and other approaches offers tremendous potential in the field of point-of-care medical diagnosis. While Raman imaging has focused on both in vivo and ex vivo approaches, the complementary ability of IR spectroscopy lies in to screening a field of cells from a blood smear or tissue section based on the cells intrinsic molecular phenotype. This represents a potentially significant step forward compared to conventional hemotoxylin and eosin (H&E) and immune-chemical staining methods. Early studies combining FT-IR mapping and multivariate data analysis showed the potential of the technique to identify cancerous cell types in tissue sections.88,89 Complementary to Raman imaging to investigate breast90,91 and fiber optic devices to detect cervical cancer,92 IR analyses can be coupled multimodally using multivariate data analysis as we have recently shown.93 The advent of quantum cascade laser (QCL) microscopes has enabled rapid discrete wavenumber imaging of biological materials.94,95 This has been particular useful in rapid imaging of large tissue sections and applications to colorectal,96 colon,97 and breast98 cancer. In this section, we highlight some recent advances and applications in vibrational imaging modalities applied to point-of-care diagnosis from biofluids to single cells and tissues. While we focus on applications of IR imaging, we also present complementary examples in Raman imaging to illustrate the areas where each may be applied.
Infrared Imaging of Biofluids
Spectroscopy is eminently suited to the analysis of biofluids. Infrared bands can be related to different constituents of the metabolome, proteome, and lipidome of biofluids such as urine,99 whole blood,100 saliva,101 or cerebrospinal fluid (CSF).102 The acquisition of infrared spectra can be performed in a few minutes without the need for complex sample, making this technique a suitable tool for the point-of-care (POC) analysis. Due to the linearity of concentration and infrared absorbance bands (Beer–Lambert law), the technique can be used for quantification of clinical parameters103 in biological liquid104 and gas samples.105 In addition, by comparing spectra from control and infected/treated sets, spectral biomarkers can be obtained for the investigation and diagnoses of disease using a fast and simple portable infrared spectrometer. The challenge remains in extracting the important spectral marker bands from other sources of variation.106 The use of imaging techniques for the analysis of biofluids is somewhat limited in the POC context. Hitherto, the preferred choice is more cost effective and compact single point attenuated total reflection (ATR) or transmission devices, which measure the bulk of the sample and do not provide any spatial resolution. Nevertheless, researchers have been investigating recently the integration of imaging as supporting tools in the POC analysis. Hughes et al.107 applied FT-IR imaging for the assessment of the reproducibility of dry films of biofluids using transmission and ATR measurements. Their results indicated that the bulk measurements of dry films of serum are affected by inconsistencies in the deposition and drying of the sample. The authors demonstrated that FT-IR imaging was a suitable technique for studying these depositions,. They also found that the irreproducibility was sample dependent. It was shown that the 10–100 KDa extract had an irregular distribution with cracks at the center of the film. This can contribute to undesirable effects such as scattering and a lack in consistency throughout the dried film. In contrast, films obtained from the >100 KDa extract showed a uniform shape and presented less irregularities.
One of the most important sources of unevenness in dry films is the so-called ‘‘coffee ring effect’’.108 When a liquid dries, a ring-like deposit forms around the center of the drop.109 Molecules with different molecular masses and polarity migrate to the periphery at different ratios, such that the composition of the periphery and the center may be different. Recently, Choi et al.110 have investigated the dynamic process that lead to ring formation using bovine gamma globulin and bovine serum albumin. They used infrared hyperspectral images and proposed an energy-based kinetic model. Results indicated that there are differences between the dynamics of the evaporation globular and non-globular proteins, provoking segregation patterns in the edge of the dry film. Finally, the use of infrared imaging as a tool for fast serum analysis has also been recently explored by Hughes et al.111 Whereas the imaging configuration cannot compete with ATR and transmission bulk analysis in terms of compatibility, the authors investigated the use of hyperspectral images as a high throughput method for acquiring the spectra of several samples from a single image. Because the aim of the work was to provide a fast POC, discrete frequencies IR (DF-IR) using a QCL laser was employed. This enabled the acquisition of different frequencies of light, making possible to perform diagnostic models using just a few variables, which reduces considerably the acquisition time. They used a microdispenser to create dry films deposition of approximately 130 µm in diameter. In total, 56 different serum samples were deposited on a sample substrate and could be measured in a 2 × 2 mosaic image. Although, the topography of the deposition was irregular, the results reported a relative good set of standard deviation values of 0.6, 5.1 and 15.0% for datasets containing 199, 14, and nine variables, respectively. In summary, the use of vibrational spectroscopy imaging techniques in the POC analysis it is not ideal because of the size and cost of the current instruments. However, the introduction of new technologies such as DF-IR based on QCL may allow in the future the fast analysis of biofluids in the POC context.
FT-IR Imaging Applied to Single Cell Analysis for Point-of-Care Analysis
Fourier transform infrared imaging has not only been successfully applied to analyze tissue sections, but also for single cell analysis. However, this application appears more challenging due to the limited spatial resolution of this technique. This drawback can be overcome by application of a synchrotron light source.112 Although not suited to POC this methodology has been applied for the single cell analysis of Plasmodium falciparum infected red blood cells. The different stages of the Plasmodium life cycle could be discriminated by their IR absorbance in the CH-stretching region (3100–2800 cm−1) using PCA.113 In a later study, an FPA-FT-IR instrument with a thermal light source was used for the detection of red blood cells infected with P. falciparum in the trophozoite stage.114 The same setup was used in a multimodal approach in combination with Raman imaging to locate the parasite in the red blood cell, based on characteristic spectral features, such as an axial carboxyl ligand band from hemozoin at 1211 cm−1.115 Another approach to overcome the limited spatial resolution of IR imaging is the combination with atomic force microscopy (AFM), which has been applied for single cell analysis of P. falciparum infected red blood cells116 and bacteria.117,118 While this approach is technically challenging for POC it does provide a method to obtain diagnostically useful subcellular information that could be used for prognostic indicators.
Recently FT-IR imaging has been applied for stem cell research.119–123. The unmet clinical need addressed at the core of this research was the quality control of stem cells injected for regenerative medicine practice, with the danger that unidentified pluripotent cells present in the injected aliquot of cells could form into cancerous growths in the body.124 Conventional approaches for the discrimination of pluripotent cells from their differentiated progeny are either destructive or require the addition of fluorescence labeled antibodies to cell surface receptors, which compromise cell viability, promoting interest in new spectroscopic approaches that do not rely on contrast agents. Infrared imaging was used on cell monolayers produced by cell centrifugation, with the pixels size set to approximate the size of individual stem cells and quality control routines used to reject spectra where cells were clumped or missing, resulting in the acquisition of spectra that could be treated as if they were from single cells (Figure 5).120 Both human embryonic and reprogramed stem cells differentiated to early commitment states could be discriminated from their pluripotent progenitor cells with excellent sensitivity and sensitivity using this approach, based on changes in spectral bands from lipids, proteins, carbohydrates and nucleic acids. The discrimination ability of spectroscopy for early differentiation changes in the stem cells was confirmed by changes in gene expression that was observed before registration of any change in cell surface markers.124–127 Similarly, changes in the differentiation states of multipotent cells have also been reported. For example, mesenchymal stem cells differentiated towards a chondrogenic lineage could be discriminated based on changes in amide III band (1255–1215 cm−1) and C–O stretching band (1200–900 cm−1) absorbances resulting from the formation of collagen and aggrecan.114
Figure 5.
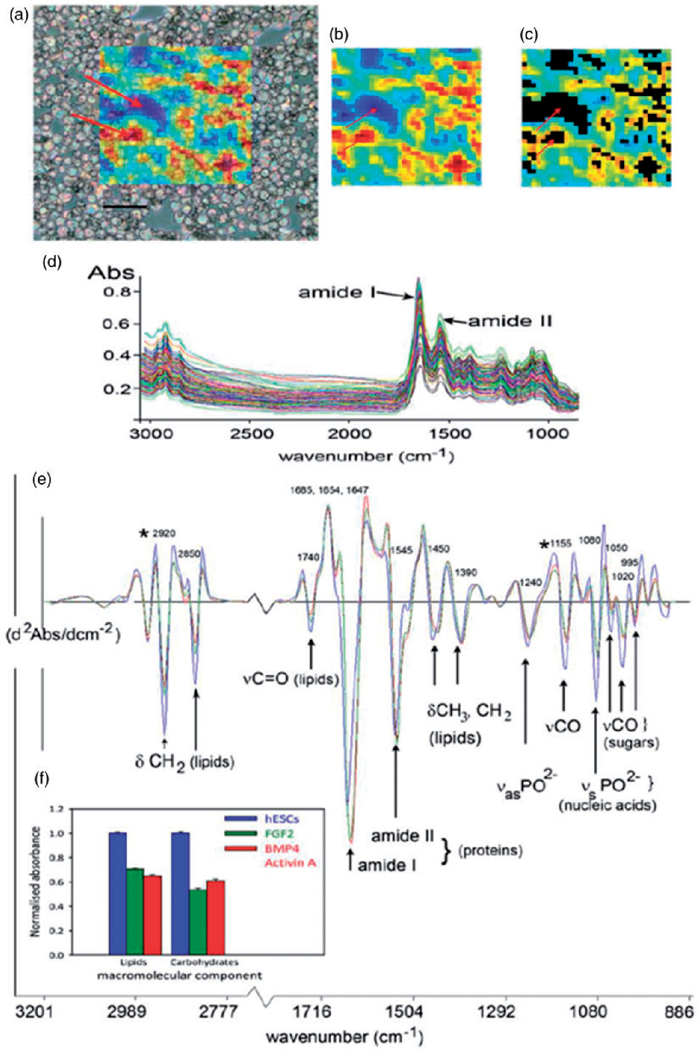
Analysis of undifferentiated and differentiated human embryonic stem cells (hESCs) using IR imaging. (a) hESCs cytospun onto a MirrIR coated slide, overlayed with a colored grid showing the area of the slide imaged by the FPA. Each colored pixel represents the area of the slide (11 µm × 11 µm projection onto sample plane) where a single FT-IR spectrum was acquired. The color scale indicates the absorbance of the amide I protein band, used as an indication of total spectral absorbance by the sample. Arrows indicate areas of low absorbance (blue) where there were no cells, or where cells overlapped, and where absorbance is high (red). Scale bar, 100 µm. (b) The same FPA image grid as shown in (a) at full optical opacity. (c) Spectral quality testing rejects spectra that are too high or low in absorbance. Black areas indicate where spectra have been rejected from the dataset, including those areas indicated by the arrows in (a). (d) The spectral data set for hESCs in one experiment (n = 192), following quality testing, but prior to spectral pre-processing. The two prominent amide I and II protein bands at ~ 1650 and 1540 cm−1, respectively, are indicated. (e) Average spectra from the experiment in (d), for hESCs (n = 192), cells treated with cytokines BMP4/Act A for four days (n = 137), and those with the cytokine FGF for four days (n = 132). Prominent bands in the spectra have been assigned to functional group vibrations and corresponding macromolecular classes. (f) Histograms showing mean integrated areas for prominent lipid and glycogen bands (asterisked at 2920 and 1155 cm−1 in panel (e) in normalized second derivative spectra from the experiment in (d). The difference between the means of areas for both bands was significantly different between hESCs and differentiated progeny in this experiment (p < 0.001, by ANOVA). Error bars indicate standard errors of the means (hESC, n = 192; BMP4/Act A, n = 137; FGF2, n = 132).
All this work with stem cells involved dried cellular deposits posing a barrier to translation of IR spectroscopic methods for the quality control of stem cells used in regenerative medicine therapies. Since water causes a strong IR background, the investigation of living cells in their natural environment using IR imaging represents a challenge. Here, microfluidic approaches that control the thickness of the water layer might provide remedy.128,129 Moreover, the water penetration issue by IR light may be overcome using Quantum Cascade Laser infrared sources, which can even penetrate skin to distances up to 50–100 µm, opening up the possibility of live cell imaging in the near future.130
FT-IR Imaging for Point-of-Care Diagnosis of Tissue Sections
Fourier transform infrared (FT-IR) imaging has been applied in a diagnostic context for a variety of tissues based on the biochemical fingerprint of the cells that constitute the tissue section. Studies in this space have focused on biopsy tissues with no papers applying the technique directly on patients. The potential of the technology to look at histological sections of excised tissue has been exploited by utilizing multivariate methods to generate false-colour maps based on spectral similarity. Characterization of skin sections or mineralized tissue such as bone has been described. Bone quality was for example assessed by FT-IR and Raman imaging by Kimura-Suda et al., by observing the PO43− band between 1200–900 cm−1, which is affected by changes in the calcium phosphate composition and indicates bone maturity.131 In another study, the collagen quality in bones was established by the investigation of IR bands around 1770–1570 and 1250–1100 cm−1 that correlate to osseous non-enzymatic cross-links.132 Moreover, FT-IR imaging in combination with partial least squares regression (PLSR) has been shown to be able to predict the collagen and elastin content in aorta membranes and the collagen maturity by the ratio of IR band intensities at 1660 and 1690 cm−1, this way prognosticating the risk for abdominal aortic aneurisms.133
Cancer causes one in six deaths worldwide, however, improved management including quick and easy screening tools could help preventing millions of deaths each year.134 Cancerous tissue can be identified from normal using FT-IR imaging methods. FT-IR imaging was for example used in several studies for the diagnosis of breast cancer.135–138 To overcome long measurement times, several recent studies focus on the optimization of this method based on only a few discrete frequencies.139,140 A study by Verdonck et al.138 implements the microenvironment of breast cancer cells by discriminating between tumor and normal phenotypes of breast endothelial cells that are either adjacent or remotely located to the tumor. While tumor tissue could be discriminated from normal tissue based on amide I and amide II bands and nucleic acid phosphate vibrations around 1210–1050 cm−1 using principal component analysis (PCA), adjacent and remote tissue was distinguished by PLS-DA based on bands at 1450 and 1000 cm−1.138
Multimodal Vibrational Imaging
There is now a wide range of available techniques for the chemical imaging of biological samples, including X-ray fluorescence (XRF), mass spectrometry imaging (MSI), laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) and vibrational techniques such as infrared and Raman spectroscopy. Each technique focuses on a different part of the cell such as the metallome, proteome, lipidome and metabolome, such that each provides only a part of the picture. Hence, samples can be measured by more than one modality, providing hopefully a more holistic picture of the cellular phenotype. Recently, there have been attempts to integrate Infrared and Raman images from red blood cells and algae in an extended hyperspectral image where every pixel contains both infrared and Raman spectra. Correlation of information obtained by the different techniques through data fusion has been demonstrated to provide more information than the sum of the parts, with information obtained by the infrared and Raman improving band assignments.88 Figure 6 shows the cluster map (Figure 6a) of a Micrasterias algal cell (see visible image on Figure 6c). Figure 6b and d show the average spectra from both infrared and Raman spectrometers. The correlation of modalities was especially useful when investigating lipids and carbohydrates. They show similar bands corresponding to C–H and C–O stretching bonds, but lipids are generally more active in Raman than carbohydrates, and this fact can be used to distinguish between the two images of the compounds. Another example of the integration of infrared and Raman imaging was presented by Lash et al.141 In this study the authors combined both vibrational modalities with MALDI-TOF to study hamster brain tissue. The analysis of the three modalities using two-dimensional correlation spectroscopy (2D-COS) enabled the investigation of auto and heterospectral correlations, which assisted the band assignment.
Figure 6.
UHCA of the multimodal image of a single Micrasterias algal cell. (a) Custer image. (b) Average infrared spectra of each class. (c) Visible image. (d) Raman average spectra of each class. Reproduced with permission from Perez-Guaita et al.93 Copyright 2017 Elsevier.
Raman Imaging of Tissue
Raman spectroscopy-based imaging of tissues has been broadly applied in the context of spectral histopathology, with tissue alterations identified on the basis of molecular changes, in a noninvasive and histopathologist-independent matter. Multiple examples of ex vivo RS-based detection of tissue abnormalities can be found in the literature, including the use of conventional Raman spectroscopy,142 as well SERS,142,143 Stimulated Raman spectroscopy (SRS),144,145 and coherent anti-Stokes Raman spectroscopy (CARS).146 In particular, the narrowband coherent techniques, such as CARS and SRS offer the ability for high speed imaging, with video rate acquisition times.144–146 RS-based spectral histopathology has proven to be useful for a great variety of tissue pathologies ranging from cancer45,143 or fibrosis,147 to detection of heterotopic ossification,148 and even the presence of malaria parasites in the spleen.149 As Raman spectroscopy is applicable for live, unfixed, and hydrated samples, prompting more and more attempts towards adaptation of RS-based spectral histopathology in vivo, in the form of Raman-assisted surgeries. This is of particular importance for the case of tumor resections, as they remain the main approach in cancer therapy, with the outcome directly depending on the ability for precise and complete removal of malignant tissue. The potential for RS-based precise determination of tumor margins in real time was shown first by Haka et al. using Raman spectra collected in vivo, during partial mastectomy.43 Since conventional Raman spectroscopy has a depth limitation, an approach based on spatially offset Raman spectroscopy (SORS), enabling to probe deep tissue (0.5–2 mm below the surface), was shown to have high sensitivity (95%) and specificity (100%) for breast tumor margin assessment.44 Kircher et al. demonstrated a the capability of SERS-based tumor margin determination performed in vivo, in a murine model during resection.150 More recently, an intraoperative application of a RS-based, label-free cancer detection system for human glioma was shown.151 St-Arnaud et al. developed a non-contact mesoscopic Raman imaging probe with a wide field of view (3.5 × 4.0 mm), enabling hyperspectral imaging of soft tissues ex vivo, with a potential for assisted-surgery application.152 The well proven ex vivo ability of RS based imaging to detect various tissue abnormalities, matching (and often exceeding) the capabilities of gold standard histopathology, together with recent rapid technological developments aimed at real-time in vivo applications create a strong base for practical, intraoperative use of RS imaging in clinics in the future.
Raman Imaging for Single Cell Analysis
Raman spectroscopy is continuously becoming more popular in the context of potential clinical applications for single cell analysis. This is primarily due to the fact that it offers the ability to obtain biochemical information from living cells in a label-free, noninvasive manner.153 The technique requires no fixation or desiccation opening up a broad range of possibilities for in vivo applications. However, the fact that spontaneous Raman scattering is a week effect (only approximately 1 × 108 of photons are scattered non-elastically154) results in long acquisition time and/or need for usage of relatively high power, unfavorable for biological materials.153,154 The long data acquisition time together with lack of standardized, robust and reliable protocols for user-friendly data analysis are the main factors currently limiting the transition of RS to the clinics.153 Nevertheless, in the recent decades multiple improvements in the instrumentation and data analysis software have largely overcome these limitations. The applications of single cell Raman imaging, particularly from the last decade, are becoming closer to the real clinical implementation.155–166 RS-based single cell imaging is especially widely utilized in cancer related studies.155–159 The ability of RS to differentiate between normal as cancerous cells has been demonstrated.155 More recently, surface enhanced Raman spectroscopy (SERS) was used for imaging cancer markers as well as for cell discrimination.156,157 The SERS technique is gaining particular popularity as it provides a substantial enhancement of signal, thus increasing the sensitivity and enabling faster data acquisition times.156–159 The approach has been applied for in vivo cancer detection, although with focus on the single spectra rather than imaging.158 MacLaughlin et al. demonstrated a SERS-based approach for detection of malignant B cells, by simultaneous use of three nanoparticle tags as labels of surface proteins, showing the potential of SERS imaging to detect leukaemia. Nanoparticle-mediated Raman imaging of cells, aimed at POC diagnosis, was recently demonstrated by Cho et al., who combined Raman active nanoprobes (RAN) with microfluidic chip to process blood samples and identify circulating cancer stem cells as well as tumor cells.159
Aside from cancer applications, RS-based diagnostic approach is continuously explored with respect to other diseases. Since Wood et al. demonstrated the ability to detect Plasmodium falciparum directly in infected red blood cells,160 several researchers have utilized Raman spectroscopy for malaria studies.161,162 In particular, the ability of resonance Raman spectroscopy combined with multivariate data analysis for fast parasite detection was shown, indication also the potential of this approach to identify low-pigmented forms (such as the early trophozoite stage) (Figure 7).166 Recently, Brückner et al. demonstrated a new Raman imaging setup enabling an automated hemozoin detection in early ring stages of Plasmodium falciparum, with the acquisition time of 10 s.165
Figure 7.
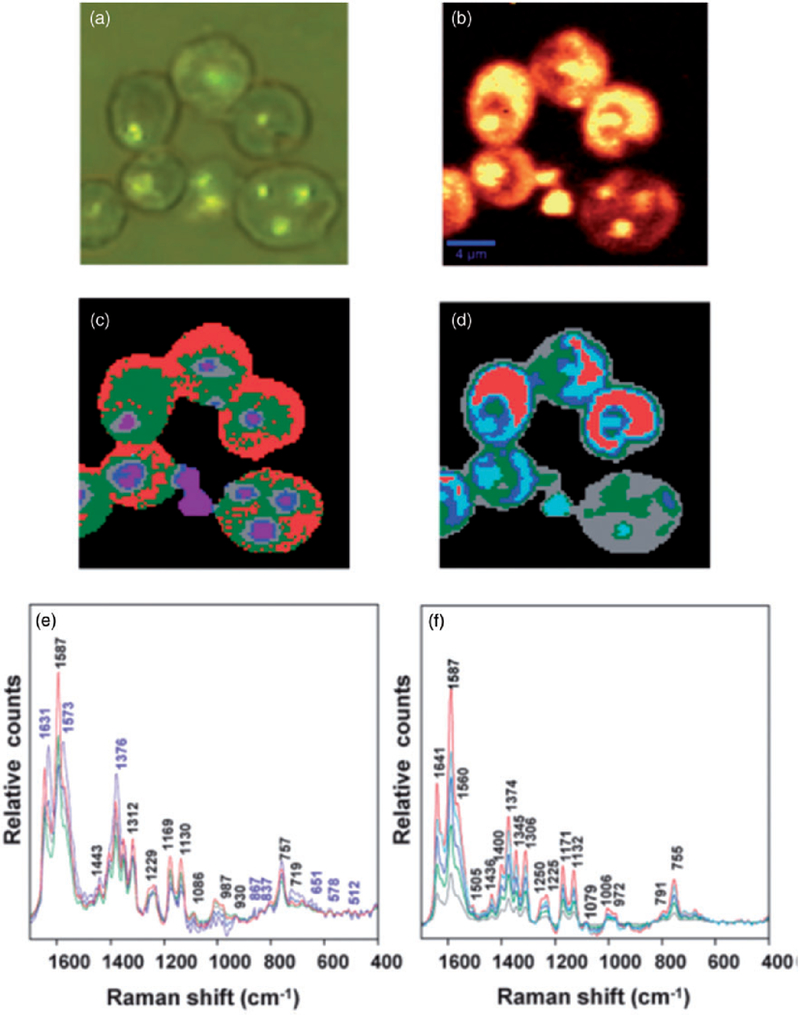
(a) Visible micrograph of infected RBCs demonstrating the partial darkfield effect, visualizing haemozoin deposits. (b) Chemical map generated by integrating the region between 1680 and 1620 cm−1. (c) Map of distribution of classes obtained using unsupervised hierarchical cluster analysis (UHCA), using the D-values distance algorithm for the 1700–1300 cm−1 range for five clusters. (c) UHCA map generated using the Euclidean distance algorithm for the 1700–1300 cm−1 range for five clusters. (e) Mean spectra corresponding to classes presented in (d). The purple labels correspond to bands mainly associated with haemozoin while the black labels are characteristic hemoglobin bands. (f) Mean spectra corresponding to classes presented in (e). The spectra show characteristic bands of hemoglobin, but not haemozoin.
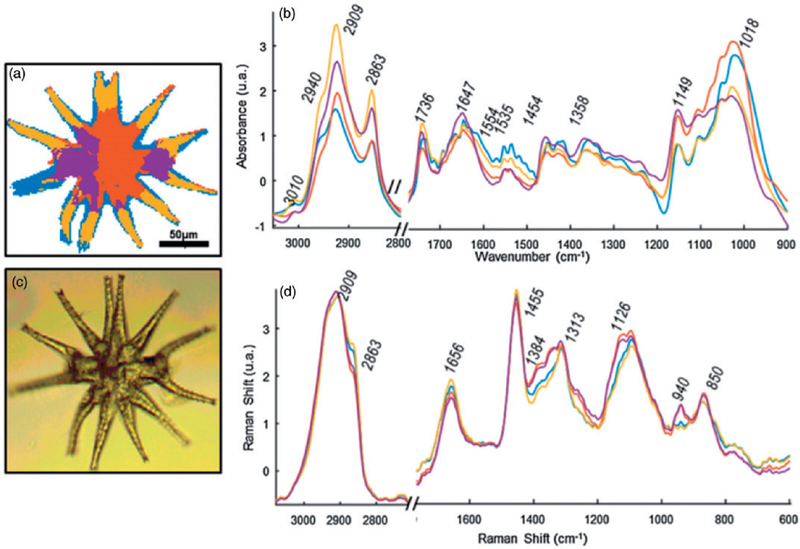
The ability to probe the macromolecular architecture of living oocytes using 3D Raman imaging was demonstrated by Heraud et al.,164 opening the way towards noninvasive comprehensive oocyte phenotyping (Figure 8). The research demonstrated the advantages of imaging living cells as opposed to fixed ones, showing both morphological changes in the three dimensional architecture as well as chemical changes when the oocytes were aldehyde fixed. Indeed, an in vivo oocyte marker band was identified observed only in living cells (Figure 8). The work leads the way towards assessment of oocyte competency in the context of assisted reproductive technologies.
Figure 8.
A photomicrograph of (a) functional and (b) fixed oocytes investigated with the use of air objective (100 x/0.90 NA) in the MII stages; Integration Raman maps of a specific bands were obtained with 532 nm laser wavelength and with a sampling density of 1 µm (maximal spatial resolution equal to 0.33 µm); K-means clustering (KMC) results with the eight main classes were presented with average spectrum for each class. In (a) we have additionally presented the zoom-in of the spectral region which corresponds to the ‘‘band of life’’27 for the single spectra extracted from the nucleic acids class. The Raman intensities in the region of 300–1900 cm−1 were scaled by factor of two comparing to CH-stretching region and lower region below 300 cm−1. A spectral class corresponding to substrate signal observed surrounding the oocytes was removed from the image (black pixels).
Summary and Outlook for IR Imaging Measurements
The application of FT-IR, QCL, and Raman imaging at point-of-care is still in its infancy. While technological advancements have been made, clinicians and pathologists must drive the translation of the technology to the clinic. It is only through their collaboration and ability to get ethical clearance to enable clinical trials of the technology that can be applied to patients presenting at hospitals and medical centers. It is clear that the images generated by these technologies are diagnostically useful but the challenge remains to batch process tissue sections and analyze the enormous data sets in a timely manner. The approach works best if a simple question is been addressed like collagen versus no collagen in FT-IR images where there are distinct marker bands that can be easily discerned from other tissue types. Different tissue densities, substrate contributions, tissue processing artifacts can lead to erroneous modeling especially when the difference between the molecular phenotype of the cells are similar. In FT-IR imaging, the closest to point-of-care translation is the Diem group who have been applying the technology to discriminate benign from malignant lung tumours.167,168 The detailed images show the potential of the technique to identify individual cell types. More recently Bhargava et al.169 demonstrated accurate subtyping from molecular properties of epithelial cells and the microenvironment using high-definition Fourier transform infrared spectroscopic imaging combined with machine learning algorithms. The advent of Raman 2D imaging in light-sheet microscopy offers the potential for 2D and 3D imaging of tissues and cells in rapid time170 and combination IR imaging systems are commercially just around the corner with the that can achieve sub-micron spatial resolution IR imaging and spectroscopy. For example, the commercial Mirage system works on the principle of photothermal IR spectroscopy (PT-IR) providing transmission quality FT-IR spectra, even in reflection mode and thus sample thickness is longer an issue. The system may soon be coupled to a Raman system providing the first fully integrated infrared/Raman imaging system ideal for cell and tissue point-of-care imaging.
Data Analysis and Understanding
Use of any spectroscopy technique for rapid and accessible decision making relies on two critical aspects: First, the data must be consistent, reproducible, and quantitative with the disease state. Second, algorithms that interpret the data to provide recognition of a physiologic state must be robust and provide rapid assessment. While sampling for homogeneous liquids and films is well established, here we focus on rapid analysis from complex, morphologically heterogeneous materials, discuss how recorded data may be affected by the sampling geometry, and show how fundamental progress in spectroscopy is leading to better understanding of such effects. We then summarize available mathematical methods that provide powerful capabilities for relating recorded data to physiologic conditions. We emphasize the interplay between understanding recorded data and using it. We anticipate that this will be a core area of study for spectroscopists focused on the fundamentals, yet a critical area of understanding needed for applied spectroscopists. We review the two areas and emphasize recent developments next.
Fundamental Theory
The underlying material property that we estimate from an absorbance measurement is the refractive index. The refractive index of an absorbing sample is a complex number and the real and imaginary parts of this complex number each have physical significance. The real part of the refractive index of a material determines how light propagates through the medium (most famously, by attenuating the speed of light). In comparison, the imaginary part determines how light is attenuated in the medium, which is proportional to the measured absorbance. IR spectroscopy of optically homogeneous samples has traditionally been performed by passing light through them (in transmission sampling geometry) and measuring the attenuation. Beer’s Law171 then predicts concentration readily from the recorded absorption spectrum. This paradigm has been the central tenet of analysis in IR spectroscopy and forms the basis of most analyses.
When a sample consists of multiple chemical constituents that are spatially distributed, maps of chemical composition are more useful and local application of Beer’s law for every pixel offers a sensitivity due to the spatial localization as well as a specificity for any changes observed. Ideally, we want to find the absorption spectrum at each point on a sample and infer chemical composition from absorbance. Such a map is most commonly obtained by coupling a microscope to an FT-IR or discrete frequency IR (DF-IR) spectrometer, resulting in IR spectroscopic imaging. However, it is now being increasingly recognized that that the absorption spectra measured in samples consisting of a spatial distribution of multiple chemical constituents is different from the spectra of the individual chemicals in a spatially homogenous sample. While tissues are naturally heterogeneous,172–174 even the spectral analysis of serum or fluid samples is now being measured in microscopy formats since deposited samples are heterogeneous. Understanding spectroscopic imaging data requires us to understand the causes of such spectral differences. This, in turn, requires an understanding of light–matter interaction at the sample as well as the data collection mechanism.
Spectra of Chemically Diverse Samples.
The problem of finding accurate absorption spectra of samples with multiple chemical constituents is identical to finding the refractive index variation across the sample at different wavenumbers, i.e., the refractive index profile at different spatial locations. However, there is a complication. Whenever there is a variation in the imaginary refractive index, there is usually a corresponding change in the real part of the refractive index. This variation in real refractive index causes scattering. When light scatters, the light distribution measured at the detector is changed. A reduction in measured intensity can be misinterpreted as being due to absorption although light is just redirected due to scattering. Since the real refractive index has a wavenumber dependence, scattering is also wavenumber dependent and the measured spectra are different from what spectroscopists would expect from the absorbance of individual chemical constituents. The profile of scattered light also depends on the optics, especially the image formation lens. Put simply, the measured spectrum not only depends on intrinsic absorbance, but also on the shape of an object as well as the optics used. In order to understand these effects and consequent spectral changes further, we need a detailed model of light–matter interaction at the sample. Note that the real and imaginary parts of refractive index are not independent variables. They are connected through the well-known Kramers–Kronig relations.175 Using this relation, it might be possible to uncouple absorbance from the real refractive index of a substance, i.e., the inverse problem. The first step in this process is to model and predict the measured spectra given the refractive index distribution of a sample, i.e., the forward problem.
Methods to Predict and Mitigate Sample Effects
There are several models to understand light–matter interaction depending on the nature of light and assumptions about the nature of the sample. Two models that are useful in spectroscopic imaging of organic materials are discussed here. Our goal in modeling light–matter interaction in the mid-IR is to understand the relation between the morphology within an object and its spectrum. Samples where such effects are seen and well recognized range from simple geometric objects like spheres to multiphase polymer samples176 to single cells177,178 and complicated tissues.179
Spectra of Layered Samples.
A good model for many organic samples is to think of them as consisting of multiple layers of known thicknesses and different refractive indices (Figure 9). We first assume that the layers are homogeneous180 and then generalize by making the layers heterogeneous.181–183 This model is useful in understanding slices of tissue placed on a BaF2 slide, for example. When the sample consists of multiple homogenous layers, we first trace the propagation of one plane wave as it travels through different layers of the sample. We do so by setting up the boundary conditions at each layer using Fresnel equations184 and solving jointly for the electric field in each layer. When a focused light beam is incident on this layered sample, we decompose the focused field into its constituent plane waves (incident at different angles) through an angular spectrum decomposition.185 We simply add the electric field contributions at each point due to the different plane waves to obtain the light distribution in a sample due to a focused beam of light. Light exiting the last boundary is what we measure. Our simulations give us this output light distribution and therefore predict our measurement. Note that the refractive index of each layer can be complex. We perform these simulations at each spectral wavenumber and can predict the full measured spectrum.
Figure 9.
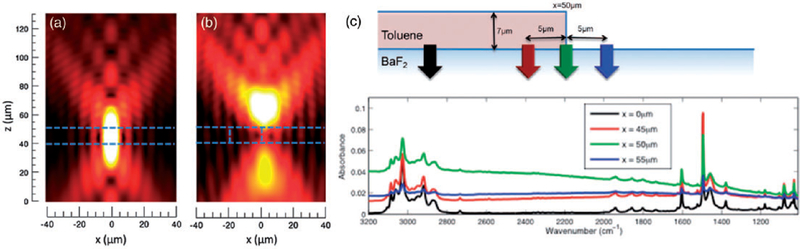
(a) The electric field distribution in a focused light beam incident of a layered sample. (b) The field is distorted significantly at the edge of a layered sample. The edge is indicated by the vertical, blue dashed line.186 (c) Spectral distortions due to edge effects is presented. When light is focused onto an edge, there is significant baseline variation as indicated by the green spectrum. This edge effect reduces as one moves away from the edge as the indicated by the black spectrum.182
When the sample consists of one or more heterogenous layers, we use coupled wave theory181 to solve for the boundary conditions. The fundamental idea behind this approach is to decompose a heterogenous layer into its Fourier series. Crudely speaking, each layer is thought of as a combination of sinusoidal ‘‘optical gratings’’ of different lines per mm i.e., ‘‘frequency’’. We know that a grating deflects light proportional to its frequency. A combination of different gratings deflects one plane wave into multiple plane waves traveling in different directions. This happens at each layer. We setup up the boundary conditions for each of these waves just as with the homogenous layer case. We solve the electric and magnetic field jointly to find out the distribution of light inside the sample. Light exiting the last layer reaches the detector and gives our measurement. This is done at different wavenumbers for the same sample and, thus, a spectrum is obtained for a sample that not only absorbs light but also scatters it. Absorption spectra are distorted in predictable ways depending on the shape of the sample and its refractive index. In layered samples, there are two important effects. The thickness of the sample results in a sinusoidal baseline variation in the sample that depends on sample thickness. The larger the thickness relative to the light’s wavelength, the larger182 this distortion. Light scattering is especially prominent at the edge of a sample. Figure 3 shows how the focused light field is distorted at the edge of a layered sample. Consequently, there is a large change in the baseline as the green spectrum in this figure indicates. Moreover, such baseline changes result in shifts in spectral peaks, which can 2.5 cm−1 or larger181 in some cases. Mitigation of these effects can be conducted by using several approaches, as discussed later.
Spectra of Spherical Objects.
Another useful model for understanding spectral distortions in heterogenous samples is to express the sample as a combination of multiple spherical objects each consisting of a single chemical constituent. Such approximations are useful in modeling cell nuclei, for example. In order to predict spectra using this model, we calculate the effects of one plane wave on a sphere whose complex refractive index is known. This can be done using classical Mie theory.187,188 In our implementation for exact understanding of the effect of spherical geometry, we again decompose the incident light into a sum of plane waves, find the electric field emitted from the sample for each plane wave and add the contributions of each incident plane wave to find the final output field. These calculations are repeated at a series of wavenumbers in order to obtain spectral data.
Figure 10 shows spectral distortions due to scattering from spherical objects. The imaginary refractive index (red) shown in Figure 10a is the ideal spectrum that we would expect from PMMA. However, when we measure a spectrum from PMMA spheres of 2.5 µm and 5 µm radii, the spectra have significant distortions that are caused by scattering. These effects can be large, but predictable as shown in Figure 10b. In order to arrive at the ‘‘true’’ absorbance spectrum, there are a few iterative algorithms187,189 that have been proposed. These involve first getting an estimate of the sphere radius, subtracting a baseline from data based on our prediction model and iterating till the prediction model and data converge. These methods work well if the assumptions of the model, especially regarding the spherical nature of the sample, corresponds to the experiment. A spherical model for scattering is used extensively as a model for single pixels from biological materials. Other shapes, for example, cylindrical objects have also been analyzed190 as models for fibers or fungi. An interesting result is that as a collection of spheres increasingly fills volume, the spectra converge to a ‘‘steady state’’ spectrum, alluding to an effective medium model.187,191–193 Together, a vast majority of samples can be modeled as spheres, collections of spheres or cylinders offering a menu of approaches to model real life objects.
Figure 10.
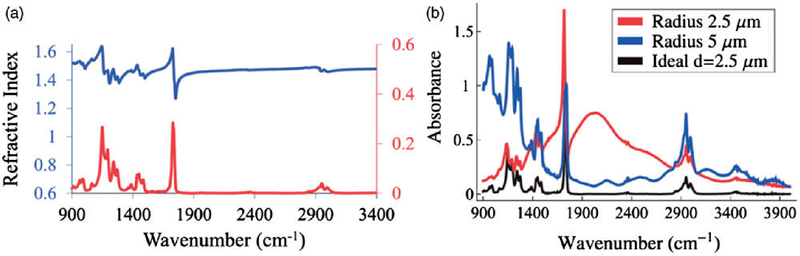
(a) The real and imaginary parts of the refractive index of PMMA are shown. (b) Spectral distortions from PMMA spheres of two different radii are presented along with the ‘‘ideal’’ spectrum (in black), which would be expected if PMMA was not spherical.189
Modeling of Instrumentation and Image Formation
While the above discussion focused on the effects of the sample, another important aspect of understanding spectral data from spectroscopic imaging is the relation between light exiting the sample focal plane and that recorded by the detector. This process involves light propagation through the entire instrument. A complete model of an instrument in order to understand the relation between the sample and data has been presented.194 This approach does not require any adjustable parameters and the rigorous approach can be used for simulation of different optical configurations. Here, light is propagated as an electromagnetic wave through the components of the optical system. Each optical component is represented by an operator that modifies the electric field. We build the system through a concatenation of operators. An advantage of this modeling approach is that there is a one-to-one correspondence between building the theory and building the instrument. Replacing one optical component in the instrument by another is the same as replacing the corresponding operator by another with the rest of the components being unchanged. This approach makes it simple to understand how data changes when an optical component is changed. A key insight from this study194 was that it was possible to obtain higher resolution spectroscopic images from the commercial instruments at that time by making small changes. Moreover, the model suggests that there are several ways of realizing this improvement. One of the simplest schemes involves replacing optical components in the path of light between the sample and detector by more appropriately designed ones. Modeling and increased availability of different designs has now led to a proliferation in the types of lenses and optical configurations186,195–200 used for imaging.
Image Formation.
Mid-IR light has a large wavelength range and the highest possible resolution that can be obtained from an FT-IR imaging instrument is determined by the diffraction limit.175 However, an appropriate design of the microscope is important in order to realize diffraction-limited resolution. Digitization and sampling of the image plays a key role in achieving diffraction limited resolution and the effective resolution obtained by a digital microscope is not only constrained by the wavenumber and numerical aperture (NA), but also by the digitization parameters of the detector array. This is determined by the design parameters of the optics between the sample and detector array. Using our instrument model, we create plots of design parameters for varying, pixel size, collector NA and wavenumber. An optimal design point is chosen based on the pixel size and NA for the highest wavenumber of interest in terms of spectroscopy. Another consequence of this modeling exercise is an understanding of the difference between FT-IR imaging (incoherent light) and imaging using QCLs (coherent light). The imaging system transfer functions are different in the two cases as detailed in the paper. Moreover, this results in strong fringes at the edges of samples in QCL based imaging systems. Such fringing artifacts are evident in measured data201 and schemes to reduce these fringes have been proposed.202 With newer designs, exceptional quality imaging at high spatial detail over the entire spectral region or the fingerprint region is now becoming common.203–210 As the number of optical configurations increase that combine novel means of illumination, light collection and detection, the role of modeling is likely to become more important. We emphasize strongly that understanding optical design for image formation is critical, but so is understanding the effect each design may have on the recorded spectrum. This is a somewhat underdeveloped area in the field of spectroscopy but offers rich opportunity and critical need as subtle changes in the spectra are increasing analyzed by complex mathematical algorithms and instruments with exceptional performance to measure very small changes are becoming increasingly available 169,211–213
Chemometric Techniques for Point-of-Care Applications
The quantification and classification of constituents in a sample can be particularly challenging given the molecular complexity seen in biological samples. In addition, the spatial heterogeneity of tissue samples ensures that all pixels are composed of an indefinite number of mixed spectral components. Finally, tissue samples are highly heterogeneous, exhibiting tremendous variation in total molecular concentration. Thus, the spectrum from a biological sample is generally treated as a ‘‘fingerprint’’ rather than as a specific molecular spectrum. While the Beer–Lambert law is still invoked to determine quantitative contributions, there is no guarantee that the total concentration or path length is constant for all pixels. The spectral histology community has generally elected to ignore path length as a variable, since the variability of fluid or tissue preparation results in the number of molecules encountered correlating poorly with underlying composition. It is therefore generally accepted to work with spectral ratios, rather than relying on raw spectra. The selection of these spectral ratios varies with application, however normalization to the Amide I protein absorbance (~1650 cm−1) is common as this is often the dominant band in biological spectra.203 This modification enforces a uniform concentration at each pixel. Another common technique is to use derivatives that are less sensitive to changes in intensities, while others yet use band shapes and ratios of carefully curated features.
Traditional Chemometric Techniques
Infrared spectra acquired from biomedical samples take on several properties that make the selection of individual molecular bands difficult. First, tissue samples are spatially diverse with morphological units varying from nanoscale to larger, resulting in pixels composed of a mixture of distinct molecular components that subtly vary across the image. Second, biological spectra exhibit a similar overall shape, making variations across a sample subtle and difficult to manually determine. For example, consider a set of spectra collected from a diverse variety of unrelated tissue types (FIGURE). Note that these spectra exhibit a common overall shape, rather similar to a Matrigel mixture,214 composed of a gelatinous protein commonly used as a substrate for cellular culture. The spectral components necessary to differentiate between cell types are subtle when compared to this dominant signal, requiring computational methods to identify and extract. Assuming that the absorption spectrum at each pixel forms a linear combination of constituent spectra, the segmentation of individual biological components is often seen as a blind source separation problem.215,216 This type of problem can be tackled with a variety of methods, with several common techniques described in this section.
Principal component analysis (PCA) is one of the most basic feature extraction techniques, taking advantage of the variance of features within a population of biological spectra , where N is the number of spectra and B the number of band measurements. This method identifies the orthogonal spectral components that vary from the mean spectrum µ, which characterizes a common biological absorbance signal (FIGURE). A set of loading vectors sorted in order of variance. A score can then be calculated for a spectrum x:
| (1) |
in which every value in s specifies the proportion of the corresponding orthogonal component from L is present in x. Since the variance is calculated globally and the loading vectors are orthogonal, it is highly unlikely that PCA captures unique or useful spectral features within L. However, if we are willing to specify a threshold such that any loading vector accounting for less than ϵ of the total variance is noise, PCA provides us with a useful tool for noise removal and dimension reduction.217 While many commercial instrumentation and software packages utilize PCA for noise removal and dimension reduction, selection of the threshold ϵ is largely subjective. While most publications select ϵ based on the percentage of the variance captured in L, this does not differentiate between signal or noise. In addition, a loading vector that captures less variance than the noise does not make that loading useless. This method remains one of the most common means of analyzing spectra, especially taking advantage of the information content of the full spectrum, but also being susceptible to artifacts arising from changes in optical configurations and sampling discussed previously. Further, interpretation of predictive signals is difficult.
Independent component analysis (ICA) provides a more robust approach for identifying spectral components by searching for independent components embedded within X. This method identifies a set of independent components that are optimized for linear independence, such that the concentration c of each component can be identified by a basis projection:
| (2) |
While the use of ICA is generally accepted in the spectroscopic community, its strength lies in separating a relatively small number of independent signals embedded in a high-dimensional (spectral) space. However, ICA relies on two underlying assumptions: first, each spectrum is a weighted linear sum of individual component spectra and, second, individual component spectra are statistically independent. The second feature is not generally satisfied in hyperspectral imaging modalities218 and is particularly uncertain in biological samples, where a discrete number of components is difficult to characterize.
Clustering methods are some of the more common unsupervised approaches for identifying biological subtypes within a sample include hierarchical and k-means clustering. In both cases, the goal is to identify a set of clusters containing spectra that minimize within-cluster variance. The k-means problem requires that the number of clusters k be known a priori. However, most applications expect to over-segment the image, incorporating a final supervised step of cluster merging. Hierarchical clustering219 offers a more comprehensive option that can be performed without a priori assumptions on the number of clusters. HCA performs an exhaustive search, often providing high-quality results for biomedical images89,220 at multiple scales.221 However this requires an exhaustive search that is often too computationally complex for large images. An alternative to both approaches that is used in the remote sensing community is a Gaussian mixture model (GMM). This method can be used in both supervised and unsupervised modes, and provides a probabilistic posterior membership value that is often more desirable for downstream image analysis.
Vertex component analysis (VCA) is a more recent technique specifically designed for hyperspectral images. Both PCA and ICA assume that the constituent spectra are statistically independent. An alternative approach is to define every spectrum in terms of a barycentric coordinate system within a simplex bounded by component spectra.222 This method allows for statistically dependent component spectra, however the resulting simplex (1) is computationally complex to calculate and (2) requires that the number of constituent components—specifying the number of edges in the simplex—be known a priori. VCA addresses the issue of computational complexity by calculating the optimal simplex using a greedy approach.218 This approach is useful because it can offer a reasonable reconstruction of component spectra as well as concentrations per pixel.223,224
Supervised Approaches
In a complex pathology with several chemical constituents changing spatially due to cell type change as well as chemical changes within cells, such as a tumor biopsy (Figure 11), unsupervised approaches may be susceptible to finding changes that arise from either factor or a complicated convolution of spectral and spatial factors. The information desired for care, however, is well defined. For inputs into care, the cellular composition and discrete disease states are typically needed and these are well-defined clinically. Supervised learning methods allow for more robust classification of samples into these types of classes (tissue cell identity and disease) by allowing a user to submit annotated example spectra associated with known types. These techniques are more robust if the user has a priori understanding of how the sample is structured, particularly when pixels exhibiting distinct spectra are known to be associated with the same tissue type (ex. cytoplasm versus nuclei).225
Figure 11.
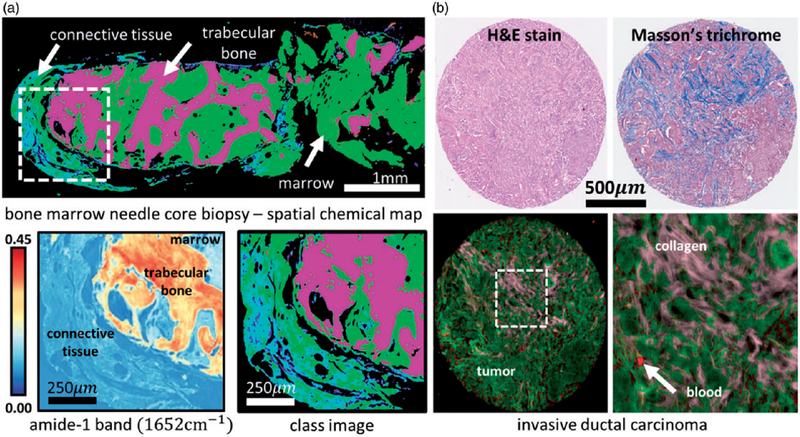
Bone and breast cancer histology classified using unsupervised and supervised machine learning. (a) Histological biopsies showing bone marrow fibrosis are shown, including a raw intensity map of the Amide I band and the resulting k-means classification results (k = 4). (b) Breast tumor biopsies (invasive ductal carcinoma) from a tissue microarray classified using a Bayesian classifier. Cancer relevant tissue types (epithelium, collagen, blood) were labeled in normal biopsies, and even simple classification methods can characterize tissues with high spatial variations, which is a common trait for tumor biopsies.
Naïve Bayes classification is one of the simplest supervised methods for placing spectra into discrete categories. This method relies on a set of representative samples to establish a probability distribution for each feature, such as an individual wavelength or PCA score. Bayesian classifiers can be trained quickly for preliminary results, and were some of the first techniques used for classification of tissue biopsies imaged using FT-IR microscopy.226,227 The major limitation for this type of classifier is often characterized as its assumption that all input features are independent. While this assumption is rarely true, these classifiers are routinely used for preliminary classification results owing to their speed and simplicity.
Random forests (RFs) are one of the more robust techniques for classifying spectra, and are often applied to unprocessed data. This technique relies on randomly sampling spectral features and using these to generate a set of decision trees that vote on the correct classification. The key advantage of an RF is its ability to readily represent multimodal distribution, which can commonly occur if a single class contains multiple chemical components. In addition, training can be performed rapidly because of the Monte Carlo approach used to sample the spectrum. These methods can be applied quickly to large data sets without dimension reduction.204,228–230 The major disadvantage of an RF is the resulting classifier size, which is dependent on the training set size and modality of the data. For biological samples, high-performance classifiers can require gigabytes of space.
Support vector machines (SVMs) are common in hyperspectral classification. SVMs attempt to draw an optimal boundary separating classes by maximizing its distance from neighboring training samples. While the traditional SVM approach use a linear boundary, the common use of a kernel trick to compute dot products in high-dimensional spaces provide a more robust, and computationally intensive, algorithm for nonlinear boundaries. SVMs utilizing a radial basis function (RBF) kernel231 are considered one of the highest-performing classification algorithms currently available for hyperspectral data.
Emerging Machine Learning Approaches
Pattern recognition is an extremely active area of research, particularly with the more recent exploration of deep learning methods. Hyperspectral imaging in general, and biomedical spectroscopy in particular, can readily benefit from recent research in several areas. In this section, we will discuss several potential applications in both feature selection and classification. While classification has always been a prominent goal in biomedical spectroscopy, the area of feature selection is becoming more important with the availability of discrete frequency imaging techniques232 that can dramatically reduce image acquisition time into the realm of clinical feasibility.
Feature selection reduces a data set by selecting a critical subset of features optimal for classification. In contrast to feature extraction techniques, such as PCA, ICA, and VCA, feature selection uses an untransformed subset of the original data. In the case of biomedical spectroscopy, these are individual wavelengths or can be combinations of wavelengths. Careful feature selection and use of limited features in predictions actually enabled the first large scale study in tissue classification with IR imaging.226 Recent work on feature selection,233 and dimension reduction in general, has focused on taking advantage of sparsity.234 In the area of vibrational spectroscopy, it is well known that bands contain redundant information. The individual bands between 1600–1700 cm−1 describing amide I are highly correlated. This certainly does not imply that the bands are completely redundant–in fact a significant body of work is focused on deconvolving information within this region. However, the additional information encoded within this region may not be necessary to solve a desired classification problem. One of the most basic sparsity-based feature selection algorithms is the least absolute shrinkage and selection operator (LASSO).235 LASSO is in a class of ℓ1 optimization problems that integrate the minimization of the absolute sum of the coefficients into the cost function:
| (3) |
where X is the matrix of training spectra, Y is the desired output value for the associated spectrum, β is the coefficient or weight of each feature, and ϵ is an input parameter that defines the sparsity of the desired feature set. LASSO forms the basis for many common feature selection methods in hyperspectral imaging,236 with promising applications in biomedical infrared spectroscopy.237 The two major disadvantages of this method with respect to hyperspectral imaging are (1) its limitation to linear fitting, which is often too simple for complex biomedical samples, and (2) the somewhat arbitrary selection of a single feature from a highly correlated group.
Alternative methods include minimum redundancy maximum relevance (mRMR)238 feature selection, which focuses on selecting features by using a greedy approach. The major limitation here lies in the use of the greedy algorithm, which starts with a single optimal feature that correlates well with the desired classification variable and iteratively adds features until the desired number is selected. The general disadvantage to of greedy approaches is that they can get caught in local minima, particularly with high-dimensional data sets such as those produced using FT-IR imaging. Some of the most promising approaches incorporate random sampling and Monte Carlo techniques. Randomized methods, such as genetic algorithms,140,205 provide three major advantages over systematic and greedy approaches. First, they reduce the chance of getting caught in local minima by relying on random sampling in combination with optimization, allowing them to be used with more complex cost functions. Second, Monte Carlo methods are well known to scale favorably to high-dimensional data. Third, randomized techniques are often highly parallel, allowing them to be readily optimized on future data parallel hardware architectures. The major cost of these techniques comes in the form of computational horsepower. However, the availability of highly parallel consumer-grade hardware such as graphics processing units (GPUs) may remove these constraints in the near future. Deep learning methods239,240 have been an extremely active area of research, and their performance on image classification and identification tasks has been extraordinary.241 In particular, deep convolutional neural networks (CNNs)242 provide a structured method for extracting spatial features using large training sets. CNNs operate by training a set of correlational filter banks that are applied iteratively to an image, allowing the hierarchical extraction of spatial features. These features become increasingly abstract, and are ultimately input into a traditional classifier, most frequently a fully connected artificial neural network (ANN) (Figure 12). While CNNs have seen increasing use in remote sensing,243–245 their current use in vibrational spectroscopy has been limited.246
Figure 12.
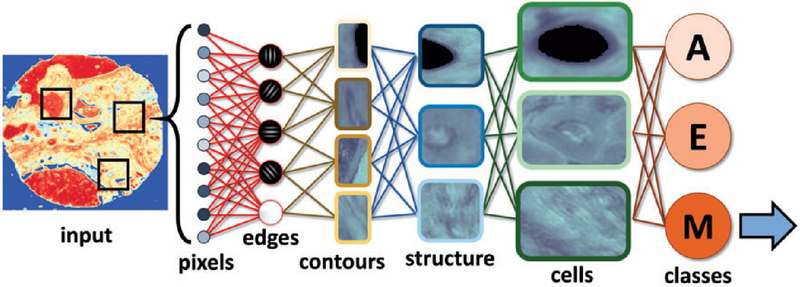
Convolutional neural networks decompose spatial features into hierarchical structures for classification. A small spatial region (including spectra) is selected as input. The network applies a series of pre-learned convolutional filters to identify a hierarchy of spatial features. The final layer is used for classification, providing a posterior probability for any cell type. Once the spatial features are learned, they can alternatively be used as input to more traditional classifiers, such as support vector machines
Rapid Computing for Spectral Analysis
Several biomedical applications, such as tissue histology, collect spatially resolved spectral images at resolutions comparable to traditional histology. Commercially available infrared imaging systems can achieve pixel sizes less than 2 µm, amounting to four million spectra in a 1 mm × 1 mm tissue microarray (TMA) core. A spectral resolution of 8 cm−1 in the range of 800–4000 cm2 and 32-bit floating point precision would result in over 6 GB of data. An image of an entire histology slide would require over 3 TB. Managing data at this magnitude requires a specialized toolset, while new imaging techniques can be used to reduce data size. These are obviously critical to developing techniques that can be used at the point of care. It is interesting to note that computing and storage capabilities have grown considerably over the past two decades, enabling faster and better analyses but also increasing the infrastructure need to deliver results in a robust manner for point of care applications.
Reducing Acquired and Analyzed Data
Recent research instruments and commercial imaging systems utilize tunable quantum cascade lasers (QCLs)95,247,248 which provide significantly higher signal strength for larger focal plane array (FPA) detectors, improving acquisition time of hyperspectral data sets.206 In addition, these instruments allow selective discrete frequency infrared (DF-IR) imaging at specified wavelengths, significantly reducing acquisition time and data size. The DF-IR approach provides the potential for clinical applicability at the point of care, provided that the appropriate wavelengths are known a priori. In practice, one would expect the appropriate wavelengths to be selected for optimal clinical viability (diagnosis, prognosis, prediction, etc.). For complex biological samples, this will likely require full spectral imaging to identify the necessary bands using feature selection.140 Recent advances in compressive sensing, such as single-pixel imaging,249 also offer opportunities to speed image acquisition. These approaches have been used in other imaging domains250 and are beginning to find application in hyperspectral image acquisition.251 We believe that a major direction for spectroscopists will lie in using their expertise to understand biological spectra, assess which data need to be acquired with the interpretation of classification algorithms and help design instruments that optimally record the data for routine applications in care.
Multiprocessor Approaches and Software
Another increasingly useful array of tools are software applications that can manage and process terabyte-scale hyperspectral images. Current tools integrated into Matlab and Python are limited by memory size. The most common commercial tools include ENVI and IDL (Harris Geospatial), which provide algorithms that are optimized for asynchronous processing and data streaming from secondary storage. Programming toolkits such as SIproc252 provide open-source alternatives, but minimal user interfaces often hinder adoption by the broader community but provide the potential for integration into other excellent data mining toolkits such as Orange.253 Custom software that integrates instrumentation, protocols needed for application(s) and visualization for interpretation by non-spectroscopists is also likely to be a major direction.
Visualization and Integration with Clinical Knowledge
Visualization plays a key role in translating hyperspectral data into a format useful for clinicians. Most existing publications rely on color-mapping classes and clusters alongside adjacent stained histology. Interactive methods may provide some assistance254 to end-users. Other alternatives involve digital staining, using machine learning to directly map hyperspectral images to patterns histologists are currently trained to interpret.255 However, many classifiers provide information that cannot be captured using these methods, such as posterior probabilities of class membership and estimated concentration percentages. Visualization of processed hyperspectral data is likely a rich area for future research. While spectroscopists focused on spectral properties of individual molecules to establish the fundamentals of the field of study, application for point of care in biological domains will require an understanding of both the origins and limits of accuracy of the signal related to biomedical knowledge as well as a deep integration with analysis and visualization tools.
Summary and Outlook for IR Imaging Measurements
A fundamental understanding of the effect of sampling and measurement configurations on the spectra of samples is a strongly emerging and highly relevant area. Together with biomedical understanding, effective development of practical protocols for point of care analysis can be developed. The advances in artificial intelligence and computational hardware open new opportunities for spectroscopy to be applied to problems in human health, with a special focus on leveraging the high dimensional analytic capability of emerging methods as well as advances in hardware to achieve near real-time analyses that are needed for point of care applications.
Conclusion
Point of care determinations from vibrational spectroscopy have shown great promise. Several trends become clear from studies conducted thus far. First, the application of vibrational spectroscopy can alter the standard of care by providing rapid and accurate assessments. By providing new information and reducing the number of steps or time required for analyses, the value offered by vibrational spectroscopic techniques is now well demonstrated. Second, the need is to develop economical and practical technology from the proof of concept studies as well as partnerships with users such that the techniques can be translated. The use of computer algorithms can greatly aid in this direction by providing quantitative information in an easy to use format (images or predictive numbers indicating risk). Finally, the need for high quality data for rapid assessments is driving spectroscopists to a greater understanding of the origins of signals recorded and their limitations. This interplay between fundamental developments and practical technology for new applications will provide higher performance, cost effective designs and computation integrated tools that vastly outperform those in use at the point of care as well as those used by spectroscopists.
Acknowledgments
Financial support of the EU, the ‘‘Thüringer Ministerium für Wirtschaft, Wissenschaft und Digitale Gesellschaft’’, the ‘‘Thüringer Aufbaubank’’, the Federal Ministry of Education and Research, Germany (BMBF), the German Science Foundation, the ‘‘Fonds der Chemischen Industrie’’ and the Carl-Zeiss Foundation is greatly acknowledged. Furthermore, we would like to thank the Leibniz-Gemeinschaft for financial support of the research alliance ,,Leibniz Health Technologies’’. Financial support was also provided by the National Institutes of Health via grants R01CA197516, R01 GM117594, and R01EB009745.
References
- 1.Stone N, Kendall C, Smith J, Crow P, et al. ‘‘Raman Spectroscopy for Identification of Epithelial Cancers’’. Faraday Discuss 2004. 126: 141–157. [DOI] [PubMed] [Google Scholar]
- 2.Kendall C, Stone N, Shepherd N, Geboes K, et al. ‘‘Raman Spectroscopy, a Potential Tool for the Objective Identification and Classification of Neoplasia in Barrett’s Oesophagus’’. J. Pathol 2003. 200(5): 602–609. [DOI] [PubMed] [Google Scholar]
- 3.Dochow S, Latka I, Becker M, Spittel R, et al. ‘‘Multicore Fiber with Integrated Fiber Bragg Gratings for Background-Free Raman Sensing’’. Opt. Express 2012. 20(18): 20156–20169. [DOI] [PubMed] [Google Scholar]
- 4.Santos LF, Wolthuis R, Koljenović S, Almeida RM, et al. ‘‘Fiber-Optic Probes for in Vivo Raman Spectroscopy in the High-Wavenumber Region’’. Anal. Chem 2005. 77(20): 6747–6752. [DOI] [PubMed] [Google Scholar]
- 5.Draga RO, Grimbergen MC, Vijverberg PL, Swol CFV, et al. ‘‘In Vivo Bladder Cancer Diagnosis by High-Volume Raman Spectroscopy’’. Anal. Chem 2010. 82(14): 5993–5999. [DOI] [PubMed] [Google Scholar]
- 6.Stevens O, Petterson IEI, Day JC, Stone N. ‘‘Developing Fibre Optic Raman Probes for Applications in Clinical Spectroscopy’’. Chem. Soc. Rev 2016. 45(7): 1919–1934. [DOI] [PubMed] [Google Scholar]
- 7.Day JC, Stone N. ‘‘A Subcutaneous Raman Needle Probe’’. Appl. Spectrosc 2013. 67(3): 349–354. [DOI] [PubMed] [Google Scholar]
- 8.Petterson IEI, Day JC, Fullwood LM, Gardner B, et al. ‘‘Characterisation of a Fibre Optic Raman Probe Within a Hypodermic Needle’’. Anal. Bioanal. Chem 2015. 407(27): 8311–8320. [DOI] [PubMed] [Google Scholar]
- 9.Matousek P, Stone N. ‘‘Development of Deep Subsurface Raman Spectroscopy for Medical Diagnosis and Disease Monitoring’’. Chem. Soc. Rev 2016. 45(7): 1794–1802. [DOI] [PubMed] [Google Scholar]
- 10.Stone N, Matousek P. ‘‘Advanced Transmission Raman Spectroscopy: A Promising Tool for Breast Disease Diagnosis’’. Cancer Res 2008. 68(11): 4424–4430. [DOI] [PubMed] [Google Scholar]
- 11.Stone N, Baker R, Rogers K, Parker AW, et al. ‘‘Subsurface Probing of Calcifications with Spatially Offset Raman Spectroscopy (SORS): Future Possibilities for the Diagnosis of Breast Cancer’’. Analyst 2007. 132(9): 899–905. [DOI] [PubMed] [Google Scholar]
- 12.Ghita A, Matousek P, Stone N. ‘‘High Sensitivity Non-Invasive Detection of Calcifications Deep Inside Biological Tissue Using Transmission Raman Spectroscopy’’. J. Biophotonics 2018. 11(1): E201600260. [DOI] [PubMed] [Google Scholar]
- 13.Matousek P, Stone N. ‘‘Prospects for the Diagnosis of Breast Cancer by Noninvasive Probing of Calcifications Using Transmission Raman Spectroscopy’’. J. Biomed. Opt 2007. 12(2): 024008. [DOI] [PubMed] [Google Scholar]
- 14.Lieber CA, Majumder SK, Ellis DL, Billheimer DD, et al. ‘‘In Vivo Nonmelanoma Skin Cancer Diagnosis Using Raman Microspectroscopy’’. Lasers Surg. Med 2008. 40(7): 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui H, Zhao J, Mclean DI, Zeng H. ‘‘Real-Time Raman Spectroscopy for in Vivo Skin Cancer Diagnosis’’. Cancer Res 2012. 72(10): 2491–500. [DOI] [PubMed] [Google Scholar]
- 16.Matousek P, Clark I, Draper E, Morris MD, et al. ‘‘Subsurface Probing in Diffusely Scattering Media Using Spatially Offset Raman Spectroscopy’’. Appl. Spectrosc 2005. 59(4): 393–400. [DOI] [PubMed] [Google Scholar]
- 17.Buckley K, Kerns JG, Gikas PD, Birch HL, et al. ‘‘Measurement of Abnormal Bone Composition in Vivo Using Noninvasive Raman Spectroscopy’’. IBMS BoneKEy 2014. 11(602): 1–3. (DOI: 10.1038/Bonekey.2014.97). [DOI] [Google Scholar]
- 18.Esmonde-White KA, Esmonde-White FW, Holmes CM, Morris MD, et al. ‘‘Alterations to Bone Mineral Composition as an Early Indication of Osteomyelitis in the Diabetic Foot’’. Diabetes Care 2013. 36(11): 3652–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim MG, Wong Kee Song LM, Marcon NE, Wilson BC. ‘‘In Vivo Near-Infrared Raman Spectroscopy: Demonstration of Feasibility During Clinical Gastrointestinal Endoscopy’’. Photochem. Photobiol 2000. 7(1): 146–150. [DOI] [PubMed] [Google Scholar]
- 20.Almond LM, Hutchings J, Lloyd G, Barr H, et al. ‘‘Endoscopic Raman Spectroscopy Enables Objective Diagnosis of Dysplasia in Barrett’s Esophagus’’. Gastrointest. Endosc 2014. 79(1): 37–45. [DOI] [PubMed] [Google Scholar]
- 21.Day J, Bennett R, Smith B, Kendall C, et al. ‘‘A Miniature Confocal Raman Probe for Endoscopic Use’’. Phys. Med. Biol 2009. 54(23): 7077. [DOI] [PubMed] [Google Scholar]
- 22.Shetty G, Kendall C, Shepherd N, Stone N, et al. ‘‘Raman Spectroscopy: Elucidation of Biochemical Changes in Carcinogenesis of Oesophagus’’. Br. J. Cancer 2006. 94(10): 1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin K, Wang J, Zheng W, Wang J, Ho KY, et al. ‘‘Rapid Fiber-Optic Raman Spectroscopy for Real-Time in Vivo Detection of Gastric Intestinal Metaplasia During Clinical Gastroscopy’’. Cancer Prev. Res 2016. 9(6): 476–483. [DOI] [PubMed] [Google Scholar]
- 24.Bergholt MS, Lin K, Wang J, Zheng W, et al. ‘‘Simultaneous Fingerprint and High-Wavenumber Fiber-Optic Raman Spectroscopy Enhances Real-Time in Vivo Diagnosis of Adenomatous Polyps During Colonoscopy’’. J. Biophotonics 2016. 9(4): 333–342. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Lin K, Zheng W, Ho KY, et al. ‘‘Comparative Study of the Endoscope-Based Bevelled and Volume Fiber-Optic Raman Probes for Optical Diagnosis of Gastric Dysplasia in Vivo at Endoscopy’’. Anal. Bioanal. Chem 2015. 407(27): 8303–8310. [DOI] [PubMed] [Google Scholar]
- 26.Stone N, Stavroulaki P, Kendall C, Birchall M, et al. ‘‘Raman Spectroscopy for Early Detection of Laryngeal Malignancy: Preliminary Results’’. Laryngoscope 2000. 110(10 Pt. 1): 1756–1763. [DOI] [PubMed] [Google Scholar]
- 27.Lin K, Zheng W, Lim CM, Huang Z. ‘‘Real-Time in Vivo Diagnosis of Laryngeal Carcinoma with Rapid Fiber-Optic Raman Spectroscopy’’. Biomed. Opt. Exp 2016. 7(9): 3705–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Short MA, Lam S, McWilliams AM, Ionescu DN, et al. ‘‘Using Laser Raman Spectroscopy to Reduce False Positives of Autofluorescence Bronchoscopies: a Pilot Study’’. J. Thoracic Oncol 2011. 6(7): 1206–1214. [DOI] [PubMed] [Google Scholar]
- 29.Santos IP, Barroso EM, Bakker Schut TC, Caspers PJ, et al. ‘‘Raman Spectroscopy for Cancer Detection and Cancer Surgery Guidance: Translation to the Clinics’’. Analyst 2017. 142(17): 3025–3047. [DOI] [PubMed] [Google Scholar]
- 30.Smith J, Kendall C, Sammon A, Christie-Brown J, et al. ‘‘Raman Spectral Mapping in the Assessment of Axillary Lymph Nodes in Breast Cancer’’. Technol. Cancer Res. Treat 2003. 2(4): 327–331. [DOI] [PubMed] [Google Scholar]
- 31.Horsnell JD, Smith JA, Sattlecker M, Sammon A, et al. ‘‘Raman Spectroscopy: A Potential New Method for Intra-Operative Assessment of Axillary Lymph Nodes’’. Surgeon 2012. 10(3): 123–127. [DOI] [PubMed] [Google Scholar]
- 32.Isabelle M, Rogers KD, Stone N. ‘‘Correlation Mapping: Rapid Method for Identification of Histological Features and Pathological Classification in Mid Infrared Spectroscopic Images of Lymph Nodes’’. J. Biomed. Opt 2010. 15(2): 026030. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd GR, Orr LE, Christie-Brown J, McCarthy K, et al. ‘‘Discrimination Between Benign, Primary and Secondary Malignancies in Lymph Nodes from the Head and Neck Utilising Raman Spectroscopy and Multivariate Analysis’’. Analyst 2013. 138(14): 3900–3908. [DOI] [PubMed] [Google Scholar]
- 34.Horsnell J, Stonelake P, Christie-Brown J, Shetty G, et al. ‘‘Raman Spectroscopy: A New Method for Intra-Operative Assessment of Axillary Lymph Nodes’’. Analyst 2010. 135(12): 3042–3047. [DOI] [PubMed] [Google Scholar]
- 35.Creager AJ, Geisinger KR. ‘‘Intraoperative Evaluation of Sentinel Lymph Nodes for Breast Carcinoma: Current Methodologies’’. Adv. Anat. Pathol 2002. 9(4): 233–243. [DOI] [PubMed] [Google Scholar]
- 36.Tew K, Irwig L, Matthews A, Crowe P, et al. ‘‘Meta-Analysis of Sentinel Node Imprint Cytology in Breast Cancer’’. Br. J. Surg 2005. 92(9): 1068–1080. [DOI] [PubMed] [Google Scholar]
- 37.Keshtgar MR. “Intra-Operative Assessment of Sentinel Nodes”. In: Sibbering M, editor. The Association of Breast Surgery at BASO Yearbook 2009. Pp. 62–65.
- 38.Blumencranz P, Whitworth P, Deck K, Rosenberg A, et al. ‘‘Scientific Impact Recognition Award. Sentinel Node Staging for Breast Cancer: Intraoperative Molecular Pathology Overcomes Conventional Histologic Sampling Errors’’. Am. J. Surg 2007. 194(4): 426–432. [DOI] [PubMed] [Google Scholar]
- 39.Tafe LJ, Schwab MC, Lefferts JA, Wells WA, et al. ‘‘A Validation Study of a New Molecular Diagnostic Assay: The Dartmouth-Hitchcock Medical Center Experience with the Genesearch BLN Assay in Breast Sentinel Lymph Nodes’’. Exp. Mol. Pathol 2010. 88(1): 1–6. [DOI] [PubMed] [Google Scholar]
- 40.Tamaki Y, Akiyama F, Iwase T, Kaneko T, et al. ‘‘Molecular Detection of Lymph Node Metastases in Breast Cancer Patients: Results of a Multicenter Trial Using the One-Step Nucleic Acid Amplification Assay’’. Clin. Cancer Res 2009. 15(8): 2879–2884. [DOI] [PubMed] [Google Scholar]
- 41.Visser M, Jiwa M, Horstman A, Brink AA, et al. ‘‘Intra-Operative Rapid Diagnostic Method Based on CK19 mRNA Expression for the Detection of Lymph Node Metastases in Breast Cancer’’. Int. J. Cancer 2008. 122(11): 2562–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haka AS, Volynskaya ZI, Gardecki JA, Nazemi J, et al. ‘‘Diagnosing Breast Cancer Using Raman Spectroscopy: Prospective Analysis’’. J. Biomed. Opt 2009. 14(5): 054023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haka AS, Volynskaya ZI, Gardecki JA, Nazemi J, et al. ‘‘In Vivo Margin Assessment During Partial Mastectomy Breast Surgery Using Raman Spectroscopy’’. Cancer Res 2006. 66(6): 3317–3322. [DOI] [PubMed] [Google Scholar]
- 44.Keller MD, Vargis E, Mahadevan-Jansen A, De Matos Granja N, et al. ‘‘Development of a Spatially Offset Raman Spectroscopy Probe for Breast Tumor Surgical Margin Evaluation’’. J. Biomed. Opt 2011. 16(7): 077006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong K, Rowlands CJ, Varma S, Perkins W, et al. ‘‘Diagnosis of Tumors During Tissue-Conserving Surgery with Integrated Autofluorescence and Raman Scattering Microscopy’’. Proc. Natl. Acad. Sci. USA 2013. 110(38): 15189–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barroso EM, Smits R, Bakker Schut T, Ten Hove I, et al. ‘‘Discrimination Between Oral Cancer and Healthy Tissue Based on Water Content Determined by Raman Spectroscopy’’. Anal. Chem 2015. 87(4): 2419–2426. [DOI] [PubMed] [Google Scholar]
- 47.Barroso EM, Smits RW, Van Lanschot CG, Caspers PJ, et al. ‘‘Water Concentration Analysis by Raman Spectroscopy to Determine the Location of the Tumor Border in Oral Cancer Surgery’’. Cancer Res 2016. 76(20): 5945–5953. [DOI] [PubMed] [Google Scholar]
- 48.Jermyn M, Mok K, Mercier J, Desroches J, et al. ‘‘Intraoperative Brain Cancer Detection with Raman Spectroscopy in Humans’’. Sci. Transl. Med 2015. 7(274): 274ra219. [DOI] [PubMed] [Google Scholar]
- 49.Vaqas B, O’Neill K, Short M, Zeng H, et al. ‘‘Optical Biopsies in Neurosurgery: Raman Spectroscopy for the Real-Time Identification of Tumours During Surgery’’. Neuro-Oncology 2016. 18(Suppl. 4): iv8. [Google Scholar]
- 50.Hollon T, Lewis S, Freudiger CW, Sunney Xie X, et al. ‘‘Improving the Accuracy of Brain Tumor Surgery via Raman-Based Technology’’. Neurosurg. Focus 2016. 40(3): E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crow P, Uff JS, Farmer JA, Wright MP, et al. ‘‘The Use of Raman Spectroscopy to Identify and Characterize Transitional Cell Carcinoma in Vitro’’. BJU Int 2004. 93(9): 1232–1236. [DOI] [PubMed] [Google Scholar]
- 52.Kendall C, Hutchings J, Barr H, Shepherd N, et al. ‘‘Exploiting the Diagnostic Potential of Biomolecular Fingerprinting with Vibrational Spectroscopy’’. Faraday Discuss 2011. 149: 279–290. [DOI] [PubMed] [Google Scholar]
- 53.Lieber CA, Nethercott HE, Kabeer MH. ‘‘Cancer Field Effects in Normal Tissues Revealed by Raman Spectroscopy’’. Biomed. Opt. Exp 2010. 1(3): 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh S, Sahu A, Deshmukh A, Chaturvedi P, et al. ‘‘In Vivo Raman Spectroscopy of Oral Buccal Mucosa: a Study on Malignancy Associated Changes (MAC)/Cancer Field Effects (CFE)’’. Analyst 2013. 138(14): 4175–4182. [DOI] [PubMed] [Google Scholar]
- 55.Sathyavathi R, Dingari NC, Barman I, Prasad P, et al. ‘‘Raman Spectroscopy Provides a Powerful, Rapid Diagnostic Tool for the Detection of Tuberculous Meningitis in Ex Vivo Cerebrospinal Fluid Samples’’. J. Biophotonics 2013. 6(8): 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klener J, Hofbauerová K, Bartoš A, Říčný J, et al. ‘‘Instability of Cerebrospinal Fluid After Delayed Storage and Repeated Freezing: A Holistic Study by Drop Coating Deposition Raman Spectroscopy’’. Clin. Chem. Lab. Med 2014. 52(5): 657–664. [DOI] [PubMed] [Google Scholar]
- 57.Kamińska A, Witkowska E, Kowalska A, Skoczyńska A, et al. ‘‘Highly Efficient SERS-Based Detection of Cerebrospinal Fluid Neopterin as a Diagnostic Marker of Bacterial Infection’’. Anal. Bioanal. Chem 2016. 408(16): 4319–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. ‘‘Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options’’. Nature Rev. Microbiol 2015. 13(5): 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brecher SM. ‘‘Complicated Urinary Tract Infections: What’s a Lab to Do?’’ J. Clin. Microbiol 2016. 54(5): 1189–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouatra S, Aziat F, Mandal R, Guo AC, et al. ‘‘The Human Urine Metabolome’’. PLoS One 2013. 8(9): E73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schröder U-C, Bokeloh F, O’Sullivan M, Glaser U, et al. ‘‘Rapid, Culture-Independent, Optical Diagnostics of Centrifugally Captured Bacteria from Urine Samples’’. Biomicrofluidics 2015. 9(4): 044118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schröder U-C, Ramoji A, Glaser U, Sachse S, et al. ‘‘Combined Dielectrophoresis–Raman Setup for the Classification of Pathogens Recovered from the Urinary Tract’’. Anal. Chem 2013. 85(22): 10717–10724. [DOI] [PubMed] [Google Scholar]
- 63.Kloss S, Kampe B, Sachse S, Rösch P, et al. ‘‘Culture Independent Raman Spectroscopic Identification of Urinary Tract Infection Pathogens: A Proof of Principle Study’’. Anal. Chem 2013. 85(20): 9610–9616. [DOI] [PubMed] [Google Scholar]
- 64.Premasiri WR, Sauer-Budge AF, Lee JC, Klapperich CM, et al. ‘‘Rapid Bacterial Diagnostics via Surface Enhanced Raman Microscopy’’. Spectroscopy 2012. 27(6): S8–31. [PMC free article] [PubMed] [Google Scholar]
- 65.Catala C, Mir-Simon B, Feng X, Cardozo C, et al. ‘‘Online SERS Quantification of Staphylococcus aureus and the Application to Diagnostics in Human Fluids’’. Adv. Mater. Technol 2016. 1(8): 1600163. [Google Scholar]
- 66.Boardman AK, Wong WS, Premasiri WR, Ziegler LD, et al. ‘‘Rapid Detection of Bacteria from Blood with Surface-Enhanced Raman Spectroscopy’’. Anal. Chem 2016. 88(16): 8026–8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ngo HT, Freedman E, Odion RA, Strobbia P, et al. ‘‘Direct Detection of Unamplified Pathogen RNA in Blood Lysate Using an Integrated Lab-in-a-Stick Device and Ultrabright SERS Nanorattles’’. Sci. Rep 2018. 8(1): 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen F, Flaherty BR, Cohen CE, Peterson DS, et al. ‘‘Direct Detection of Malaria Infected Red Blood Cells by Surface Enhanced Raman Spectroscopy’’. Nanomedicine 2016. 12: 1445–1451. [DOI] [PMC free article] [PubMed]
- 69.Kozicki M, Czepiel J, Biesiada G, Nowak P, et al. ‘‘The Ring-Stage of Plasmodium falciparum Observed in RBCs of Hospitalized Malaria Patients’’. Analyst 2015. 140(23): 8007–8016. [DOI] [PubMed] [Google Scholar]
- 70.Neugebauer U, Trenkmann S, Bocklitz T, Schmerler D, et al. ‘‘Fast Differentiation of SIRS and Sepsis from Blood Plasma of ICU Patients Using Raman Spectroscopy’’. J. Biophotonics 2014. 7(3–4): 232–240. [DOI] [PubMed] [Google Scholar]
- 71.Kamińska A, Sprynskyy M, Winkler K, Szymborski T. ‘‘Ultrasensitive SERS Immunoassay Based on Diatom Biosilica for Detection of Interleukins in Blood Plasma’’. Anal. Bioanal. Chem 2017. 409(27): 6337–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bonifacio A, Dalla Marta S, Spizzo R, Cervo S, et al. ‘‘Surface-Enhanced Raman Spectroscopy of Blood Plasma and Serum Using Ag and Au Nanoparticles: A Systematic Study’’. Anal. Bioanal. Chem 2014. 406(9–10): 2355–2365. [DOI] [PubMed] [Google Scholar]
- 73.Tatarkovič M, Synytsya A, Šťovíčková L, Bunganič B, et al. ‘‘The Minimizing of Fluorescence Background in Raman Optical Activity and Raman Spectra of Human Blood Plasma’’. Anal. Bioanal. Chem 2015. 407(5): 1335–1342. [DOI] [PubMed] [Google Scholar]
- 74.Khan S, Khurram M, Ali H, Ullah R, et al. ‘‘Evaluation of Raman Spectroscopy in Comparison to Commonly Performed Dengue Diagnostic Tests’’. J. Biomed. Opt 2016. 21(9): 095005. [DOI] [PubMed] [Google Scholar]
- 75.Khan S, Ullah R, Khan A, Wahab N, et al. ‘‘Analysis of Dengue Infection Based on Raman Spectroscopy and Support Vector Machine (SVM)’’. Biomed. Opt. Exp 2016. 7(6): 2249–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anwar S, Firdous S. ‘‘Optical Diagnostic of Hepatitis B (HBV) and C (HCV) from Human Blood Serum Using Raman Spectroscopy’’. Laser Phys. Lett 2015. 12(7): 076001. [Google Scholar]
- 77.Kloss S, Lorenz B, Dees S, Labugger I, et al. ‘‘Destruction-Free Procedure for the Isolation of Bacteria from Sputum Samples for Raman Spectroscopic Analysis’’. Anal. Bioanal. Chem 2015. 407(27): 8333–8341. [DOI] [PubMed] [Google Scholar]
- 78.Gonchukov S, Sukhinina A, Bakhmutov D, Biryukova T, et al. ‘‘Periodontitis Diagnostics Using Resonance Raman Spectroscopy on Saliva’’. Laser Phys. Lett 2013. 10(7): 075610. [Google Scholar]
- 79.Ghebremedhin M, Yesupriya S, Crane NJ. “Surface Enhanced Raman Spectroscopy as a Point-of-Care Diagnostic for Infection in Wound Effluent”. Proc. Optical Diagnostics and Sensing XVI: Toward Point-of-Care Diagnostics 2016. 9715.
- 80.Assmann C, Kirchhoff J, Beleites C, Hey J, et al. ‘‘Identification of Vancomycin Interaction with Enterococcus Faecalis Within 30 min of Interaction Time Using Raman Spectroscopy’’. Anal. Bioanal. Chem 2015. 407(27): 8343–8352. [DOI] [PubMed] [Google Scholar]
- 81.Kang WCM, Wong YL, Piotrowski M, Teo WPJ, et al. ‘‘A Gold Nanostar-Based SERS Platform for Point-of-Care Diagnostics of Carbapenemase-Producing Enterobacteriacae’’ Proc. SPIE 10340 International Conference on Biophotonics V, 2017. [Google Scholar]
- 82.Schröder UC, Kirchhoff J, Hübner U, Mayer G, et al. ‘‘On-Chip Spectroscopic Assessment of Microbial Susceptibility to Antibiotics Within 3.5 Hours’’. J. Biophotonics 2017. 10(11): 1547–1557. [DOI] [PubMed] [Google Scholar]
- 83.Kirchhoff J, Glaser U, Bohnert JA, Pletz MW, et al. ‘‘Simple Ciprofloxacin Resistance Test and Determination of Minimal Inhibitory Concentration Within 2 h Using Raman Spectroscopy’’. Anal. Chem 2018. 90(3): 1811–1818. [DOI] [PubMed] [Google Scholar]
- 84.Dekter H, Orelio C, Morsink M, Tektas S, et al. ‘‘Antimicrobial Susceptibility Testing of Gram-Positive and -Negative Bacterial Isolates Directly from Spiked Blood Culture Media with Raman Spectroscopy’’. Eur. J. Clin. Microbiol. Infect. Dis 2017. 36(1): 81–89. [DOI] [PubMed] [Google Scholar]
- 85.Premasiri WR, Chen Y, Williamson PM, Bandarage DC, et al. ‘‘Rapid Urinary Tract Infection Diagnostics by Surface-Enhanced Raman Spectroscopy (SERS): Identification and Antibiotic Susceptibilities’’. Anal. Bioanal. Chem 2017. 409(11): 3043–3054. [DOI] [PubMed] [Google Scholar]
- 86.Novelli-Rousseau A, Espagnon I, Filiputti D, Gal O, et al. ‘‘Culture-Free Antibiotic-Susceptibility Determination from Single-Bacterium Raman Spectra’’. Sci. Rep 2018. 8(1): 3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathey R, Dupoy M, Espagnon I, Leroux D, et al. ‘‘Viability of 3h Grown Bacterial Micro-Colonies After Direct Raman Identification’’. J. Microbiol. Methods 2015. 109: 67–73. [DOI] [PubMed] [Google Scholar]
- 88.Lasch P, Naumann D. ‘‘FT-IR Microspectroscopic Imaging of Human Carcinoma Thin Sections Based on Pattern Recognition Techniques’’. Cell. Mol. Biol 1998. 44(1): 189–202. [PubMed] [Google Scholar]
- 89.Wood BR, Chiriboga L, Yee H, Quinn MA, et al. ‘‘Fourier Transform Infrared (FTIR) Spectral Mapping of the Cervical Transformation Zone, and Dysplastic Squamous Epithelium’’. Gynecol. Oncol 2004. 93(1): 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henry CM. ‘‘Raman Spectra of Breast Tissue’’. Anal. Chem 1996. 68(23): 718A–719A. [DOI] [PubMed] [Google Scholar]
- 91.Schaeberle MD, Kalasinsky VF, Luke JL, Lewis EN, et al. ‘‘Raman Chemical Imaging: Histopathology of Inclusions in Human Breast Tissue’’. Anal. Chem 1996. 68(11): 1829–1833. [DOI] [PubMed] [Google Scholar]
- 92.Mahadevan-Jansen A, Mitchell MF, Ramanujam N, Utzinger U, et al. ‘‘Development of a Fiber Optic Probe to Measure NIR Raman Spectra of Cervical Tissue in Vivo’’. Photochem. Photobiol 1998. 68(3): 427–431. [PubMed] [Google Scholar]
- 93.Perez-Guaita D, Kochan K, Martin M, Andrew DW, et al. ‘‘Multimodal Vibrational Imaging of Cells’’. Vib. Spectrosc 2017. 91: 46–58. [Google Scholar]
- 94.Kimber JA, Kazarian SG. ‘‘Spectroscopic Imaging of Biomaterials and Biological Systems with FTIR Microscopy Or with Quantum Cascade Lasers’’. Anal. Bioanal. Chem 2017. 409(25): 5813–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yeh K, Kenkel S, Liu JN, Bhargava R. ‘‘Fast Infrared Chemical Imaging with a Quantum Cascade Laser’’. Anal. Chem 2015. 87(1): 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bird B, Rowlette J. ‘‘High Definition Infrared Chemical Imaging of Colorectal Tissue Using a Spero QCL Microscope’’. Analyst 2017. 142(8): 1381–1386. [DOI] [PubMed] [Google Scholar]
- 97.Kroger-Lui N, Gretz N, Haase K, Kranzlin B, et al. ‘‘Rapid Identification of Goblet Cells in Unstained Colon Thin Sections by Means of Quantum Cascade Laser-Based Infrared Microspectroscopy’’. Analyst 2015. 140(7): 2086–2092. [DOI] [PubMed] [Google Scholar]
- 98.Bassan P, Weida MJ, Rowlette J, Gardner P. ‘‘Large Scale Infrared Imaging of Tissue Micro Arrays (TMAs) Using a Tunable Quantum Cascade Laser (QCL) Based Microscope’’. Analyst 2014. 139(16): 3856–3859. [DOI] [PubMed] [Google Scholar]
- 99.Shaw RA, Kotowich S, Mantsch HH, Leroux M. ‘‘Quantitation of Protein, Creatinine, and Urea in Urine by Near-Infrared Spectroscopy’’. Clin. Biochem 1996. 29(1): 11–19. [DOI] [PubMed] [Google Scholar]
- 100.Perez-Guaita D, Ventura-Gayete J, Perez-Rambla C, Sancho-Andreu M, et al. ‘‘Protein Determination in Serum and Whole Blood by Attenuated Total Reflectance Infrared Spectroscopy’’. Anal. Bioanal. Chem 2012. 404(3): 649–656. [DOI] [PubMed] [Google Scholar]
- 101.Rodrigues LM, Magrini TD, Lima CF, Scholz J, et al. ‘‘Effect of Smoking Cessation in Saliva Compounds by FTIR Spectroscopy’’. Spectrochim. Acta, Part A 2017. 174: 124–129. [DOI] [PubMed] [Google Scholar]
- 102.Yonar D, Ocek L, Tiftikcioglu BI, Zorlu Y, et al. ‘‘Relapsing-Remitting Multiple Sclerosis Diagnosis from Cerebrospinal Fluids via Fourier Transform Infrared Spectroscopy Coupled with Multivariate Analysis’’. Sci. Rep 2018. 8(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Perez-Guaita D, Garrigues S, De La Guardia M. ‘‘Infrared-Based Quantification of Clinical Parameters’’. TRaC, Trends Anal. Chem 2014. 62: 93–105. [Google Scholar]
- 104.Hosafci G, Klein O, Oremek G, Mantele W. ‘‘Clinical Chemistry Without Reagents? an Infrared Spectroscopic Technique for Determination of Clinically Relevant Constituents of Body Fluids’’. Anal. Bioanal. Chem 2007. 387(5): 1815–1822. [DOI] [PubMed] [Google Scholar]
- 105.Perez-Guaita D, Kokoric V, Wilk A, Garrigues S, et al. ‘‘Towards the Determination of Isoprene in Human Breath Using Substrate-Integrated Hollow Waveguide Mid-Infrared Sensors’’. J. Breath Res 2014. 8(2): 026003. [DOI] [PubMed] [Google Scholar]
- 106.Diem M. ‘‘Comments on Recent Reports on Infrared Spectral Detection of Disease Markers in Blood Components’’. J. Biophotonics 11(7): E201800064. [DOI] [PubMed] [Google Scholar]
- 107.Hughes C, Brown M, Clemens G, Henderson A, et al. ‘‘Assessing the Challenges of Fourier Transform Infrared Spectroscopic Analysis of Blood Serum’’. J. Biophotonics 2014. 7(3–4): 180–188. [DOI] [PubMed] [Google Scholar]
- 108.Goodacre R, Baker MJ, Graham D, Schultz ZD, et al. ‘‘Biofluids and Other Techniques: General Discussion’’. Faraday Discuss 2016. 187: 575–601. [DOI] [PubMed] [Google Scholar]
- 109.Deegan RD, Bakajin O, Dupont TF, Huber G, et al. ‘‘Capillary Flow as the Cause of Ring Stains from Dried Liquid Drops’’. Nature 1997. 389: 827–829. [Google Scholar]
- 110.Choi S, Birarda G. ‘‘Protein Mixture Segregation at Coffee-Ring: Real-Time Imaging of Protein Ring Precipitation by FTIR Spectromicroscopy’’. J. Phys. Chem. B 2017. 121(30): 7359–7365. [DOI] [PubMed] [Google Scholar]
- 111.Hughes C, Clemens G, Bird B, Dawson T, et al. ‘‘Introducing Discrete Frequency Infrared Technology for High-Throughput Biofluid Screening’’. Sci. Rep 2016. 6: 20173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harrison JP, Berry D. ‘‘Vibrational Spectroscopy for Imaging Single Microbial Cells in Complex Biological Samples’’. Front. Microbiol 2017. 8: 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wood BR, Bambery KR, Dixon MWA, Tilley L, et al. ‘‘Diagnosing Malaria Infected Cells at the Single Cell Level Using Focal Plane Array Fourier Transform Infrared Imaging Spectroscopy’’. Analyst 2014. 139(19): 4769–4774. [DOI] [PubMed] [Google Scholar]
- 114.Perez-Guaita D, Andrew D, Heraud P, Beeson J, et al. ‘‘High Resolution FTIR Imaging Provides Automated Discrimination and Detection of Single Malaria Parasite Infected Erythrocytes on Glass’’. Faraday Discuss 2016. 187: 341–352. [DOI] [PubMed] [Google Scholar]
- 115.Perez-Guaita D, Kochan K, Martin M, Andrew DW, et al. ‘‘Multimodal Vibrational Imaging of Cells’’. Vib. Spectrosc 2017. 91: 46–58. [Google Scholar]
- 116.Perez-Guaita D, Kochan K, Batty M, Doerig C, et al. ‘‘Multispectral Atomic Force Microscopy-Infrared Nano-Imaging of Malaria Infected Red Blood Cells’’. Anal. Chem 2018. 90(5): 3140–3148. [DOI] [PubMed] [Google Scholar]
- 117.Baldassarre L, Giliberti V, Rosa A, Ortolani M, et al. ‘‘Mapping the Amide I Absorption in Single Bacteria and Mammalian Cells with Resonant Infrared Nanospectroscopy’’. Nanotechnol 2016. 7(7): 075101. [DOI] [PubMed] [Google Scholar]
- 118.Kochan K, Perez-Guaita D, Pissang J, Jiang JH, et al. ‘‘In Vivo Atomic Force Microscopy-Infrared Spectroscopy of Bacteria’’. J. R. Soc. Interface 2018. 15(140): 0180115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chonanant C, Bambery KR, Jearanaikoon N, Chio-Srichan S, et al. ‘‘Discrimination of Micromass-Induced Chondrocytes from Human Mesenchymal Stem Cells by Focal Plane Array–Fourier Transform Infrared Microspectroscopy’’. Talanta 2014. 130: 39–48. [DOI] [PubMed] [Google Scholar]
- 120.Heraud P, Ng ES, Caine S, Yu QC, et al. ‘‘Fourier Transform Infrared Microspectroscopy Identifies Early Lineage Commitment in Differentiating Human Embryonic Stem Cells’’. Stem Cell Res 2010. 4(2): 140–147. [DOI] [PubMed] [Google Scholar]
- 121.Krafft C, Salzer R, Seitz S, Ern C, Schieker M. ‘‘Differentiation of Individual Human Mesenchymal Stem Cells Probed by FTIR Microscopic Imaging’’. Analyst 2007. 132(7): 647–653. [DOI] [PubMed] [Google Scholar]
- 122.Tanthanucha W, Thumanu K, Lorthongpanich C, Parnpai R, et al. ‘‘Neural Differentiation of Mouse Embryonic Stem Cells Studied by FTIR Spectroscopy’’. J. Mol. Struct 2010. 967(1–3): 189–195. [Google Scholar]
- 123.Clemens G, Flower KR, Henderson AP, Whiting A, et al. ‘‘The Action of All-Trans-Retinoic Acid (ATRA) and Synthetic Retinoid Analogues (EC19 and EC23) on Human Pluripotent Stem Cells Differentiation Investigated Using Single Cell Infrared Microspectroscopy’’. Mol. Biosyst 2013. 9(4): 677–692. [DOI] [PubMed] [Google Scholar]
- 124.Heraud P, Tobin MJ. ‘‘The Emergence of Biospectroscopy in Stem Cell Research’’. Stem Cell Res 2009. 3(1): 12–14. [DOI] [PubMed] [Google Scholar]
- 125.Cao J, Ng ES, McNaughton D, Stanley EG, et al. ‘‘The Characterisation of Pluripotent and Multipotent Stem Cells Using Fourier Transform Infrared Microspectroscopy’’. Int. J. Mol. Sci 2013. 14(9): 17453–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cao J, Ng ES, McNaughton D, Stanley EG, et al. ‘‘Fourier Transform Infrared Microspectroscopy Reveals That Tissue Culture Conditions Affect the Macromolecular Phenotype of Human Embryonic Stem Cells’’. Analyst 2013. 138(14): 4147–4160. [DOI] [PubMed] [Google Scholar]
- 127.Heraud P, Ng ES, Caine S, Yu QC, et al. ‘‘Fourier Transform Infrared Microspectroscopy Identifies Early Lineage Commitment in Differentiating Human Embryonic Stem Cells’’. Stem Cell Res 2010. 4(2): 140–147. [DOI] [PubMed] [Google Scholar]
- 128.Loutherback K, Birarda G, Chen L, Holman HYN. ‘‘Microfluidic Approaches to Synchrotron Radiation-Based Fourier Transform Infrared (SR-FTIR) Spectral Microscopy of Living Biosystems’’. Protein Peptide Lett 2016. 23(3): 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tobin M, Puskar L, Barber R, Harvey E, et al. ‘‘FTIR Spectroscopy of Single Live Cells in Aqueous Media by Synchrotron IR Microscopy Using Microfabricated Sample Holders’’. Vib. Spectrosc 2010. 53(1): 34–38. [Google Scholar]
- 130.Schwaighofer A, Brandstetter M, Lendl B. ‘‘Quantum Cascade Lasers (QCLs) in Biomedical Spectroscopy.’’ Chem. Soc. Rev 2017. 46(19): 5903–5924. [DOI] [PubMed] [Google Scholar]
- 131.Kimura-Suda H, Ito T. ‘‘Bone Quality Characteristics Obtained by Fourier Transform Infrared and Raman Spectroscopic Imaging’’. J. Oral Biosci 2017. 59(3): 142–145. [Google Scholar]
- 132.Schmidt FN, Zimmermann EA, Campbell GM, Sroga GE, et al. ‘‘Assessment of Collagen Quality Associated with Non-Enzymatic Cross-Links in Human Bone Using Fourier-Transform Infrared Imaging’’. Bone 2017. 97: 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cheheltani R, Pichamuthu J, Rao J, Weinbaum J, et al. ‘‘Fourier Transform Infrared Spectroscopic Imaging-Derived Collagen Content and Maturity Correlates with Stress in the Aortic Wall of Abdominal Aortic Aneurysm Patients’’. Cardiovasc. Eng. Technol 2017. 8(1): 70–80. [DOI] [PubMed] [Google Scholar]
- 134.World Health Organization (WHO). Fact Sheet: Cancer http://www.who.int/news-room/fact-sheets/detail/cancer [accessed 9 July 2018].
- 135.Holton SE, Bergamaschi A, Katzenellenbogen BS, Bhargava R. ‘‘Integration of Molecular Profiling and Chemical Imaging to Elucidate Fibroblast-Microenvironment Impact on Cancer Cell Phenotype and Endocrine Resistance in Breast Cancer’’. PLoS One 2014. 9(5): E96878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kumar S, Desmedt C, Larsimont D, Sotiriou C, et al. ‘‘Change in the Microenvironment of Breast Cancer Studied by FTIR Imaging’’. Analyst 2013. 138(14): 4058–4065. [DOI] [PubMed] [Google Scholar]
- 137.Kumar S, Srinivasan A, Nikolajeff F. ‘‘Role of Infrared Spectroscopy and Imaging in Cancer Diagnosis’’. Curr. Med. Chem 2018. 25(9): 1055–1072. [DOI] [PubMed] [Google Scholar]
- 138.Verdonck M, Denayer A, Delvaux B, Garaud S, et al. ‘‘Characterization of Human Breast Cancer Tissues by Infrared Imaging’’. Analyst 2016. 141(2): 606–619. [DOI] [PubMed] [Google Scholar]
- 139.Ali MH, Rakib F, Al-Saad K, Al-Saady R, et al. ‘‘A Simple Model for Cell Type Recognition Using 2D-Correlation Analysis of FTIR Images from Breast Cancer Tissue’’. J. Mol. Struct 2018. 1163: 472–479. [Google Scholar]
- 140.Mankar R, Walsh MJ, Bhargava R, Prasad S, et al. ‘‘Selecting Optimal Features from Fourier Transform Infrared Spectroscopy for Discrete-Frequency Imaging’’. Analyst 2018. 143(5): 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lasch P, Noda I. ‘‘Two-Dimensional Correlation Spectroscopy for Multimodal Analysis of FT-IR, Raman, and MALDI-TOF MS Hyperspectral Images with Hamster Brain Tissue’’. Anal. Chem 2017. 89(9): 5008–5016. [DOI] [PubMed] [Google Scholar]
- 142.Kong K, Kendall C, Stone N, Notingher I. ‘‘Raman Spectroscopy for Medical Diagnostics: From in-Vitro Biofluid Assays to in-Vivo Cancer Detection’’. Adv. Drug. Deliv. Rev 2015. 89: 121–134. [DOI] [PubMed] [Google Scholar]
- 143.Andreou C, Kishore SA, Kircher MF. ‘‘Surface-Enhanced Raman Spectroscopy: a New Modality for Cancer Imaging’’. J. Nuclear Med 2015. 56(9): 1295–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ozeki Y, Umemura W, Otsuka Y, Satoh S, et al. ‘‘High-Speed Molecular Spectral Imaging of Tissue with Stimulated Raman Scattering’’. Nature Photonics 2012. 6: 845–851. [Google Scholar]
- 145.Lu F-K, Calligaris D, Olubiyi OI, Norton I, et al. ‘‘Label-Free Neurosurgical Pathology with Stimulated Raman Imaging’’. Cancer Res 2016. 76(12): 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Camp CH Jr, Lee YJ, Heddleston JM, Hartshorn CM, et al. ‘‘High-Speed Coherent Raman Fingerprint Imaging of Biological Tissues’’. Nature Photonics 2014. 8: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin J, Lu F, Zheng W, Huang Z, et al. ‘‘Assessment of Liver Steatosis and Fibrosis in Rats Using Integrated Coherent Anti-Stokes Raman Scattering and Multiphoton Imaging Technique’’. J. Biomed. Opt 2011. 16(11): 116024. [DOI] [PubMed] [Google Scholar]
- 148.Peterson JR, Okagbare PI, De La Rosa S, Cilwa KE, et al. ‘‘Early Detection of Burn Induced Heterotopic Ossification Using Transcutaneous Raman Spectroscopy’’. Bone 2013. 54(1): 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Frame L, Brewer J, Lee R, Faulds K, et al. ‘‘Development of a Label-Free Raman Imaging Technique for Differentiation of Malaria Parasite Infected from Non-Infected Tissue’’. Analyst 2018. 143(1): 157–163. [DOI] [PubMed] [Google Scholar]
- 150.Kircher MF, De La Zerda A, Jokerst JV, Zavaleta CL, et al. ‘‘A Brain Tumor Molecular Imaging Strategy Using a New Triple-Modality MRI-Photoacoustic-Raman Nanoparticle’’. Nature Med 2012. 18(5): 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Desroches J, Jermyn M, Pinto M, Picot F, et al. ‘‘A New Method Using Raman Spectroscopy for in Vivo Targeted Brain Cancer Tissue Biopsy’’. Sci. Rep 2018. 8(1): 1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.St-Arnaud K, Aubertin K, Strupler M, Madore WJ, et al. ‘‘Development and Characterization of a Handheld Hyperspectral Raman Imaging Probe System for Molecular Characterization of Tissue on Mesoscopic Scales’’. Med. Phys 2018. 45(1): 328–339. [DOI] [PubMed] [Google Scholar]
- 153.Smith R, Wright KL, Ashton L. ‘‘Raman Spectroscopy: An Evolving Technique for Live Cell Studies’’. Analyst 2016. 141(12): 3590–3600. [DOI] [PubMed] [Google Scholar]
- 154.Smith E, Dent G. Modern Raman Spectroscopy: a Practical Approach Chichester: John Wiley And Sons, 2013. [Google Scholar]
- 155.Chan JW, Taylor DS, Zwerdling T, Lane SM, et al. ‘‘Micro-Raman Spectroscopy Detects Individual Neoplastic and Normal Hematopoietic Cells’’. Biophys. J 2006. 90(2): 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang J, Wang Z, Zong S, Song C, et al. ‘‘Distinguishing Breast Cancer Cells Using Surface-Enhanced Raman Scattering’’. Anal. Bioanal. Chem 2012. 402(3): 1093–1100. [DOI] [PubMed] [Google Scholar]
- 157.Lee S, Chon H, Lee M, Choo J, et al. ‘‘Surface-Enhanced Raman Scattering Imaging of HER2 Cancer Markers Overexpressed in Single MCF7 Cells Using Antibody Conjugated Hollow Gold Nanospheres’’. Biosens. Bioelectron 2009. 24(7): 2260–2263. [DOI] [PubMed] [Google Scholar]
- 158.Qian X, Peng X-H, Ansari DO, Yin-Goen Q, et al. ‘‘In Vivo Tumor Targeting and Spectroscopic Detection with Surface-Enhanced Raman Nanoparticle Tags’’. Nature Biotechnol 2008. 26(1): 83. [DOI] [PubMed] [Google Scholar]
- 159.Cho H-Y, Hossain MK, Lee J-H, Han J, et al. ‘‘Selective Isolation and Noninvasive Analysis of Circulating Cancer Stem Cells Through Raman Imaging’’. Biosens. Bioelectron 2018. 102: 372–382. [DOI] [PubMed] [Google Scholar]
- 160.Wood BR, Langford SJ, Cooke BM, Glenister FK, et al. ‘‘Raman Imaging of Hemozoin Within the Food Vacuole of Plasmodium falciparum Trophozoites’’. FEBS Lett 2003. 554(3): 247–252. [DOI] [PubMed] [Google Scholar]
- 161.Chen F, Flaherty BR, Cohen CE, Peterson DS, et al. ‘‘Direct Detection of Malaria Infected Red Blood Cells by Surface Enhanced Raman Spectroscopy’’. Nanomedicine 2016. 12(6): 1445–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bonifacio A, Finaurini S, Krafft C, Parapini S, et al. ‘‘Spatial Distribution of Heme Species in Erythrocytes Infected with Plasmodium falciparum by Use of Resonance Raman Imaging and Multivariate Analysis’’. Anal. Bioanal. Chem 2008. 392(7–8): 1277–1282. [DOI] [PubMed] [Google Scholar]
- 163.McLaughlin CM, Mullaithilaga N, Yang G, Ip SY, et al. ‘‘Surface-Enhanced Raman Scattering Dye-Labeled Au Nanoparticles for Triplexed Detection of Leukemia and Lymphoma Cells and SERS Flow Cytometry’’. Langmuir 2013. 29(6): 1908–1919. [DOI] [PubMed] [Google Scholar]
- 164.Heraud P, Marzec KM, Zhang QH, Yuen WS, et al. ‘‘Label-Free in Vivo Raman Microspectroscopic Imaging of the Macromolecular Architecture of Oocytes’’. Sci. Rep 2017. 7(1): 8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brückner M, Becker K, Popp J, Frosch T. ‘‘Fiber Array Based Hyperspectral Raman Imaging for Chemical Selective Analysis of Malaria-Infected Red Blood Cells’’. Anal. Chim. Acta 2015. 894: 76–84. [DOI] [PubMed] [Google Scholar]
- 166.Wood BR, Hermelink A, Lasch P, Bambery KR, et al. ‘‘Resonance Raman Microscopy in Combination with Partial Dark-Field Microscopy Lights Up a New Path in Malaria Diagnostics’’. Analyst 2009. 134(6): 1119–1125. [DOI] [PubMed] [Google Scholar]
- 167.Diem M, Ergin A, Remiszewski S, Mu X. ‘‘PS01.31: A Reagent-Free, High Resolution Lung Cancer Diagnostic Method Based on Phenotypic Infrared Spectral Imaging: Topic: Pathology’’. J. Thorac. Oncol 2016. 11(11S): S288. [Google Scholar]
- 168.Akalin A, Mu X, Kon MA, Ergin A, et al. ‘‘Classification of Malignant and Benign Tumors of the Lung by Infrared Spectral Histopathology (SHP)’’. Lab. Invest 2015. 95(4): 406–421. [DOI] [PubMed] [Google Scholar]
- 169.Mittal S, Yeh K, Leslie LS, Kenkel S, et al. ‘‘Simultaneous Cancer and Tumor Microenvironment Subtyping Using Confocal Infrared Microscopy for All-Digital Molecular Histopathology’’. Proc. Natl. Acad. Sci. USA 2018. 115(25): E5651–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rocha-Mendoza I, Licea-Rodriguez J, Marro M, Olarte OE, et al. ‘‘Rapid Spontaneous Raman Light Sheet Microscopy Using cw-Lasers and Tunable Filters’’. Biomed. Opt. Exp 2015. 6(9): 3449–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Griffiths PR, De Haseth JA, Winefordner JD. Fourier Transform Infrared Spectrometry Hoboken, NJ: Wiley-Interscience, 2007. [Google Scholar]
- 172.Diem M, Romeo M, Boydston-White S, Miljkovic M, et al. ‘‘A Decade of Vibrational Micro-Spectroscopy of Human Cells and Tissue (1994–2004)’’. Analyst 2004. 129(10): 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lasch P, Boese M, Pacifico A, Diem M. ‘‘FT-IR Spectroscopic Investigations of Single Cells on the Subcellular Level’’. Vib. Spectrosc 2002. 28: 147–157. [Google Scholar]
- 174.Diem M, Mazur A, Lenau K, Schubert J, et al. ‘‘Molecular Pathology via IR and Raman Spectral Imaging’’. J. Biophotonics 2013. 6(11–12): 855–886. [DOI] [PubMed] [Google Scholar]
- 175.Saleh BEA, Teich MC. Fundamentals of Photonics Chichester: Wiley-Interscience,1991. [Google Scholar]
- 176.Bhargava R, Wang S-Q, Koenig JL. ‘‘FT-IR Imaging of the Interface in Multicomponent Systems Using Optical Effects Induced by Differences in Refractive Index’’. Appl. Spectrosc 1998. 52(3): 323–328. [Google Scholar]
- 177.Bassan P, Byrne HJ, Lee J, Bonnier F, et al. ‘‘Reflection Contributions to the Dispersion Artefact in FTIR Spectra of Single Biological Cells’’. Analyst 2009. 134(6): 1171–1175. [DOI] [PubMed] [Google Scholar]
- 178.Quaroni L, Zlateva T, Normand E. ‘‘Detection of Weak Absorption Changes from Molecular Events in Time-Resolved FT-IR Spectromicroscopy Measurements of Single Functional Cells’’. Anal. Chem 2011. 83(19): 7371–7380. [DOI] [PubMed] [Google Scholar]
- 179.Mohlenhoff B, Romeo M, Diem M, Wood BR. ‘‘Mie-Type Scattering and Non-Beer–Lambert Absorption Behavior of Human Cells in Infrared Microspectroscopy’’. Biophys. J 2005. 88(5): 3635–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Davis BJ, Carney PS, Bhargava R. ‘‘Theory of Midinfrared Absorption Microspectroscopy: I. Homogeneous Samples’’. Anal. Chem 2010. 82(9): 3474–3486. [DOI] [PubMed] [Google Scholar]
- 181.Davis BJ, Carney PS, Bhargava R. ‘‘Theory of Mid-Infrared Absorption Microspectroscopy: II. Heterogeneous Samples’’. Anal. Chem 2010. 82(9): 3487–3499. [DOI] [PubMed] [Google Scholar]
- 182.Reddy R, Mayerich D, Walsh M, Schulmerich M, et al. “Optimizing the Design of FT-IR Spectroscopic Imaging Instruments to Obtain Increased Spatial Resolution of Chemical Species”. 9th IEEE International Symposium on Biomedical Imaging (ISBI). Barcelona, Spain; 2–5 May 2012. [Google Scholar]
- 183.Bassan P, Lee J, Sachdeva A, Pissardini J, et al. ‘‘The Inherent Problem of Transflection-Mode Infrared Spectroscopic Microscopy and the Ramifications for Biomedical Single Point and Imaging Applications’’. Analyst 2013. 138(1): 144–157. [DOI] [PubMed] [Google Scholar]
- 184.Peatross J, Ware M. Physics of Light and Optics Provo, Utah: Brigham Young University Department of Physics, 2011. [Google Scholar]
- 185.Goodman J.
- 186.Walsh MJ, Reddy RK, Bhargava R. ‘‘Label-Free Biomedical Imaging with Mid-IR Spectroscopy’’. IEEE J. Sel. Top. Quantum Electron 2012. 18(4): 1502–1513. [Google Scholar]
- 187.Bassan P, Kohler A, Martens H, Lee J, et al. ‘‘Resonant Mie Scattering (RMieS) Correction of Infrared Spectra from Highly Scattering Biological Samples’’. Analyst 2010. 135(2): 268–277. [DOI] [PubMed] [Google Scholar]
- 188.Bambery KR, Wood BR, McNaughton D. ‘‘Resonant Mie Scattering (RMieS) Correction Applied to FTIR Images of Biological Tissue Samples’’. Analyst 2012. 137(1): 126–132. [DOI] [PubMed] [Google Scholar]
- 189.Dijk T, Van Mayerich D, Carney PS, Bhargava R. ‘‘Recovery of Absorption Spectra from Fourier Transform Infrared (FT-IR) Microspectroscopic Measurements of Intact Spheres’’. Appl. Spectrosc 2013. 67(5): 546–552. [DOI] [PubMed] [Google Scholar]
- 190.Davis BJ, Scott Carney P, Bhargava R. ‘‘Theory of Infrared Microspectroscopy for Intact Fibers’’. Anal. Chem 2011. 83(2): 525–532. [DOI] [PubMed] [Google Scholar]
- 191.Bassan P, Byrne HJ, Bonnier F, Lee J, et al. ‘‘Resonant Mie Scattering in Infrared Spectroscopy of Biological Materials—Understanding the ‘Dispersion Artefact’’’. Analyst 2009. 134(8): 1586–1593. [DOI] [PubMed] [Google Scholar]
- 192.Konevskikh T, Lukacs R, Kohler A. “An Improved Algorithm for Fast Resonant Mie Scatter Correction of Infrared Spectra of Cells and Tissues”. J. Biophotonics 11(1): E201600307. [DOI] [PubMed] [Google Scholar]
- 193.Rasskazov IL, Spegazzini N, Carney PS, et al. ‘‘Dielectric Sphere Clusters as a Model to Understand Infrared Spectroscopic Imaging Data Recorded from Complex Samples’’. Anal. Chem 2017. 89(20): 10813–10818. [DOI] [PubMed] [Google Scholar]
- 194.Reddy RK, Walsh MJ, Schulmerich MV, Carney PS, et al. ‘‘High-Definition Infrared Spectroscopic Imaging’’. Appl. Spectrosc 2013. 67(1): 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Walsh MJ, Holton SE, Kajdacsy-Balla A, Bhargava R. ‘‘Attenuated Total Reflectance Fourier-Transform Infrared Spectroscopic Imaging for Breast Histopathology’’. Vib. Spectrosc 2012. 60: 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kimber JA, Foreman L, Turner B, Rich P, et al. ‘‘FTIR Spectroscopic Imaging and Mapping with Correcting Lenses for Studies of Biological Cells and Tissues’’. Faraday Discuss 2016. 187: 69–85. [DOI] [PubMed] [Google Scholar]
- 197.Wrobel TP, Vichi A, Baranska M, Kazarian SG. ‘‘Micro-Attenuated Total Reflection Fourier Transform Infrared (Micro ATR FT-IR) Spectroscopic Imaging with Variable Angles of Incidence’’. Appl. Spectrosc 2015. 69(10): 1170–1174. [DOI] [PubMed] [Google Scholar]
- 198.Ewing AV, Clarke GS, Kazarian SG. ‘‘Attenuated Total Reflection-Fourier Transform Infrared Spectroscopic Imaging of Pharmaceuticals in Microfluidic Devices’’. Biomicrofluidics 2016. 10(2): 024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mayerich D, Van Dijk T, Walsh MJ, Schulmerich MV, et al. ‘‘On the Importance of Image Formation Optics in the Design of Infrared Spectroscopic Imaging Systems’’. Analyst 2014. 139(16): 4031–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nallala J, Lloyd GR, Hermes M, Shepherd N, et al. ‘‘Enhanced Spectral Histology in the Colon Using High-Magnification Benchtop FTIR Imaging’’. Vib. Spectrosc 2017. 91: 83–91. [Google Scholar]
- 201.Kole MR, Reddy RK, Schulmerich MV, Gelber MK, et al. ‘‘Discrete Frequency Infrared Microspectroscopy and Imaging with a Tunable Quantum Cascade Laser’’. Anal. Chem 2012. 84(23): 10366–10372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ran S, Berisha S, Mankar R, Shih W-C, et al. ‘‘Mitigating Fringing in Discrete Frequency Infrared Imaging Using Time-Delayed Integration’’. Biomed. Opt. Exp 2018. 9(2): 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Baker MJ, Trevisan J, Bassan P, Bhargava R, et al. ‘‘Using Fourier Transform IR Spectroscopy to Analyze Biological Materials’’. Nature Protoc 2014. 9(8): 1771–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Leslie LS, Wrobel TP, Mayerich D, Bindra S, et al. ‘‘High Definition Infrared Spectroscopic Imaging the Lymph Node Histopathology’’. PLoS One 2015. 10(6): E0127238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mankar R, Verma V, Walsh M, Bueso-Ramos C, et al. ‘‘Imaging and Feature Selection Using GA-FDA Algorithm for the Classification of Mid-Infrared Biomedical Images’’. Microsc. Microanal 2016. 22(S3): 1008–1009. [Google Scholar]
- 206.Kuepper C, Kallenbach-Thieltges A, Juette H, Tannapfel A, et al. ‘‘Quantum Cascade Laser-Based Infrared Microscopy for Label-Free and Automated Cancer Classification in Tissue Sections’’. Sci. Rep 2018. 8(1): 7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mattson EC, Nasse MJ, Rak M, Gough KM, et al. ‘‘Restoration and Spectral Recovery of Mid-Infrared Chemical Images’’. Anal. Chem 2012. 84(14): 6173–6180. [DOI] [PubMed] [Google Scholar]
- 208.Nasse MJ, Walsh MJ, Mattson EC, Reininger R, et al. ‘‘High-Resolution Fourier-Transform Infrared Chemical Imaging with Multiple Synchrotron Beams’’. Nat. Methods 2011. 8(5): 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tiwari S, Raman J, Reddy V, Ghetler A, et al. ‘‘Towards Translation of Discrete Frequency Infrared Spectroscopic Imaging for Digital Histopathology of Clinical Biopsy Samples’’. Anal. Chem 2016. 88(20): 10183–10190. [DOI] [PubMed] [Google Scholar]
- 210.Pilling MJ, Henderson A, Gardner P. ‘‘Quantum Cascade Laser Spectral Histopathology: Breast Cancer Diagnostics Using High Throughput Chemical Imaging’’. Anal. Chem 2017. 89(14): 7348–7355. [DOI] [PubMed] [Google Scholar]
- 211.Schwaighofer A, Alcaráz MR, Kuligowski J, Lendl B. ‘‘Recent Advancements of EC-QCL Based Mid-IR Transmission Spectroscopy of Proteins and Application to Analysis of Bovine Milk’’. Biomed. Spectrosc. Imaging 2018. 7(1–2): 35–45. [Google Scholar]
- 212.Schwaighofer A, Montemurro M, Freitag S, Kristament C, et al. ‘‘Beyond Fourier Transform Infrared Spectroscopy: External Cavity Quantum Cascade Laser-Based Mid-Infrared Transmission Spectroscopy of Proteins in the Amide I and Amide II Region’’. Anal. Chem 2018. 90(11): 7072–7079. [DOI] [PubMed] [Google Scholar]
- 213.Patel CKN, Barron-Jimenez R, Dunayevskiy I, Troccoli M. “Advances in Fast Tunable Laser Spectroscopy in the Infrared”. Proc. SPIE 10639, Micro- and Nanotechnology Sensors, Systems, and Applications X, 2018. 1063920 DOI: 10.1117/12.2306940. [DOI] [Google Scholar]
- 214.Lee J, Gazi E, Dwyer J, Brown MD, et al. ‘‘Optical Artefacts in Transflection Mode FTIR Microspectroscopic Images of Single Cells on a Biological Support: The Effect of Back-Scattering Into Collection Optics’’. Analyst 2007. 132(8): 750–755. [DOI] [PubMed] [Google Scholar]
- 215.Belouchrani A, Abed-Meraim K, Cardoso JF, Moulines E. ‘‘A Blind Source Separation Technique Using Second-Order Statistics’’. IEEE Trans. Signal Process 1997. 45(2): 434–444. [Google Scholar]
- 216.Zarzoso V, Nandi AK. “Blind Source Separation”. In: Nandi AK, Editor. Blind Estimation Using Higher-Order Statistics Boston, MA: Springer, 1999. Chap. 4, Pp. 167–252. [Google Scholar]
- 217.Hughes C, Henderson A, Kansiz M, Dorling KM, et al. ‘‘Enhanced FTIR Bench-Top Imaging of Single Biological Cells’’. Analyst 2015. 140(7): 2080–2085. [DOI] [PubMed] [Google Scholar]
- 218.Nascimento JMP, Dias JMB. ‘‘Does Independent Component Analysis Play a Role in Unmixing Hyperspectral Data?’’ IEEE Trans. Geosci. Electron 2005. 43(1): 175–187. [Google Scholar]
- 219.Johnson SC. ‘‘Hierarchical Clustering Schemes’’. Psychometrika 1967. 32(3): 241–254. [DOI] [PubMed] [Google Scholar]
- 220.Lasch P, Haensch W, Naumann D, Diem M. ‘‘Imaging of Colorectal Adenocarcinoma Using FT-IR Microspectroscopy and Cluster Analysis’’. Biochim. Biophys. Acta 2004. 1688(2): 176–186. [DOI] [PubMed] [Google Scholar]
- 221.Amenabar I, Poly S, Goikoetxea M, Nuansing W, et al. ‘‘Hyperspectral Infrared Nanoimaging of Organic Samples Based on Fourier Transform Infrared Nanospectroscopy’’. Nat. Commun 2017. 8: 14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Craig MD. ‘‘Minimum-Volume Transforms for Remotely Sensed Data’’. IEEE Trans. Geosci. Electron 1994. 32(3): 542–552. [Google Scholar]
- 223.Krafft C, Diderhoshan MA, Recknagel P, Miljkovic M, et al. ‘‘Crisp and Soft Multivariate Methods Visualize Individual Cell Nuclei in Raman Images of Liver Tissue Sections’’. Vib. Spectrosc 2011. 55(1): 90–100. [Google Scholar]
- 224.Meyer T, Bergner N, Krafft C, Akimov D, et al. ‘‘Nonlinear Microscopy, Infrared, and Raman Microspectroscopy for Brain Tumor Analysis’’. J. Biomed. Opt 2011. 16(2): 021113. [DOI] [PubMed] [Google Scholar]
- 225.Nallala J, Lloyd GR, Shepherd N, Stone N. ‘‘High-Resolution FTIR Imaging of Colon Tissues for Elucidation of Individual Cellular and Histopathological Features’’. Analyst 2016. 141(2): 630–639. [DOI] [PubMed] [Google Scholar]
- 226.Fernandez DC, Bhargava R, Hewitt SM, Levin IW. ‘‘Infrared Spectroscopic Imaging for Histopathologic Recognition’’. Nat. Biotechnol 2005. 23(4): 469. [DOI] [PubMed] [Google Scholar]
- 227.Bhargava R, Fernandez DC, Hewitt SM, Levin IW. ‘‘High Throughput Assessment of Cells and Tissues: Bayesian Classification of Spectral Metrics from Infrared Vibrational Spectroscopic Imaging Data’’. Biochim. Biophys. Acta, Biomembr 2006. 1758(7): 830–845. [DOI] [PubMed] [Google Scholar]
- 228.Kallenbach-Thieltges A, Grosserü schkamp F, Mosig A, Diem M, et al. ‘‘Immunohistochemistry, Histopathology and Infrared Spectral Histopathology of Colon Cancer Tissue Sections’’. J. Biophotonics 2013. 6(1): 88–100. [DOI] [PubMed] [Google Scholar]
- 229.Mayerich DM, Walsh M, Kadjacsy-Balla A, Mittal S, Bhargava R. ‘‘Breast Histopathology Using Random Decision Forests-Based Classification of Infrared Spectroscopic Imaging Data’’. Proc. SPIE 9041, Medical Imaging Digital Pathology 2014. 904107 DOI: 10.1117/12.2043783. [DOI] [Google Scholar]
- 230.Kuepper C, Grosserueschkamp F, Kallenbach-Thieltges A, Mosig A, et al. ‘‘Label-Free Classification of Colon Cancer Grading Using Infrared Spectral Histopathology’’. Faraday Discuss 2016. 187: 105–118. [DOI] [PubMed] [Google Scholar]
- 231.Scholkopf B, Kah-Kay S, Burges CJC, Girosi F, et al. ‘‘Comparing Support Vector Machines with Gaussian Kernels to Radial Basis Function Classifiers’’. IEEE Trans. Signal Process 1997. 45(11): 2758–2765. [Google Scholar]
- 232.Wrobel TP, Kole MR, Bhargava R. ‘‘Emerging Trends and Opportunities in Discrete-Frequency Infrared and Raman Spectroscopic Imaging’’. Spectroscopy 2016. 31(6): 28–45. [Google Scholar]
- 233.Lloyd GR, Stone N. ‘‘Method for Identification of Spectral Targets in Discrete Frequency Infrared Spectroscopy for Clinical Diagnostics’’. Appl. Spectrosc 2015. 69(9): 1066–1073. [DOI] [PubMed] [Google Scholar]
- 234.Guyon I, Gunn S, Nikravesh M, Zadeh LA. Feature Extraction: Foundations, Applications Berlin; Heidelberg: Springer-Verlag, 2008. [Google Scholar]
- 235.Tibshirani R. ‘‘Regression Shrinkage and Selection via the Lasso’’. J. R. Stat. Soc. Series B. Stat. Methodol 1996. 58(1): 267–288. [Google Scholar]
- 236.Qian Y, Ye M, Zhou J. ‘‘Hyperspectral Image Classification Based on Structured Sparse Logistic Regression and Three-Dimensional Wavelet Texture Features’’. IEEE Trans. Geosci. Electron 2013. 51(4): 2276–2291. [Google Scholar]
- 237.Gajjar K, Trevisan J, Owens G, Keating PJ, et al. ‘‘Fourier-Transform Infrared Spectroscopy Coupled with a Classification Machine for the Analysis of Blood Plasma or Serum: A Novel Diagnostic Approach for Ovarian Cancer’’. Analyst 2013. 138(14): 3917–3926. [DOI] [PubMed] [Google Scholar]
- 238.Ding C, Peng H. ‘‘Minimum Redundancy Feature Selection from Microarray Gene Expression Data’’. J. Bioinform. Comput. Biol 2005. 3(2): 185–205. [DOI] [PubMed] [Google Scholar]
- 239.Lecun Y, Bengio Y, Hinton G. ‘‘Deep Learning’’. Nature 2015. 521(7553): 436–444. [DOI] [PubMed] [Google Scholar]
- 240.Schmidhuber J. ‘‘Deep Learning in Neural Networks: An Overview’’. Neural Networks 2015. 61: 85–117. [DOI] [PubMed] [Google Scholar]
- 241.He K, Zhang X, Ren S, Sun J. “Deep Residual Learning for Image Recognition”. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (CVPR). Las Vegas, Nevada; 27–30 June 2016 10.1109/CVPR.2016.90. [DOI] [Google Scholar]
- 242.Krizhevsky A, Sutskever I, Hinton GE. ‘‘Imagenet Classification with Deep Convolutional Neural Networks’’. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in Neural Information Processing Systems 25 Red Hook, NY: Curran Associates, Inc, 2012, pp.1097–1105. [Google Scholar]
- 243.Chen Y, Lin Z, Zhao X, Wang G, et al. ‘‘Deep Learning-Based Classification of Hyperspectral Data’’. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens 2014. 7(6): 2094–2107. [Google Scholar]
- 244.Nogueira K, Penatti OAB, Dos Santos JA. ‘‘Towards Better Exploiting Convolutional Neural Networks for Remote Sensing Scene Classification’’. Pattern Recognit 2017. 61(C): 539–556. [Google Scholar]
- 245.Yu S, Jia S, Xu C. ‘‘Convolutional Neural Networks for Hyperspectral Image Classification’’. Neurocomputing 2017. 219(C): 88–98. [Google Scholar]
- 246.Acquarelli J, Van Laarhoven T, Gerretzen J, Tran TN, et al. ‘‘Convolutional Neural Networks for Vibrational Spectroscopic Data Analysis’’. Anal. Chim. Acta 2017. 954: 22–31. [DOI] [PubMed]
- 247.Bassan P, Weida MJ, Rowlette J, Gardner P. ‘‘Large Scale Infrared Imaging of Tissue Micro Arrays (TMAs) Using a Tunable Quantum Cascade Laser (QCL) Based Microscope’’. Analyst 2014. 139(16): 3856–3859. [DOI] [PubMed] [Google Scholar]
- 248.Kröger N, Egl A, Engel M, Gretz N, et al. ‘‘Quantum Cascade Laser-Based Hyperspectral Imaging of Biological Tissue’’. J. Biomed. Opt 2014. 19(11): 111607. [DOI] [PubMed] [Google Scholar]
- 249.Duarte MF, Davenport MA, Takhar D, Laska JN, et al. ‘‘Single-Pixel Imaging via Compressive Sampling’’. IEEE Signal Process. Mag 2008. 25(2): 83–91. [Google Scholar]
- 250.Chan WL, Charan K, Takhar D, Kelly KF, et al. ‘‘A Single-Pixel Terahertz Imaging System Based on Compressed Sensing’’. Appl. Phys. Lett 2008. 93(12): 121105. [Google Scholar]
- 251.Studer V, Bobin J, Chahid M, Mousavi HS, et al. ‘‘Compressive Fluorescence Microscopy for Biological and Hyperspectral Imaging’’. Proc. Natl. Acad. Sci. USA 2012. 109(26): E1679–E1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Berisha S, Chang S, Saki S, Daeinejad D, et al. ‘‘SIproc: An Open-Source Biomedical Data Processing Platform for Large Hyperspectral Images’’. Analyst 2017. 142(8): 1350–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Demšar J, Curk T, Erjavec A, Gorup Č, et al. ‘‘Orange: Data Mining Toolbox in Python’’. J. Machine Learning Res 2013. 14(1): 2349–2353. [Google Scholar]
- 254.Mayerich D, Walsh M, Schulmerich M, Bhargava R. ‘‘Real-Time Interactive Data Mining for Chemical Imaging Information: Application to Automated Histopathology’’. BMC Bioinf 2013. 14: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mayerich D, Walsh MJ, Kadjacsy-Balla A, Ray PS, et al. ‘‘Stain-Less Staining for Computed Histopathology’’. Technology 2015. 3(1): 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]


