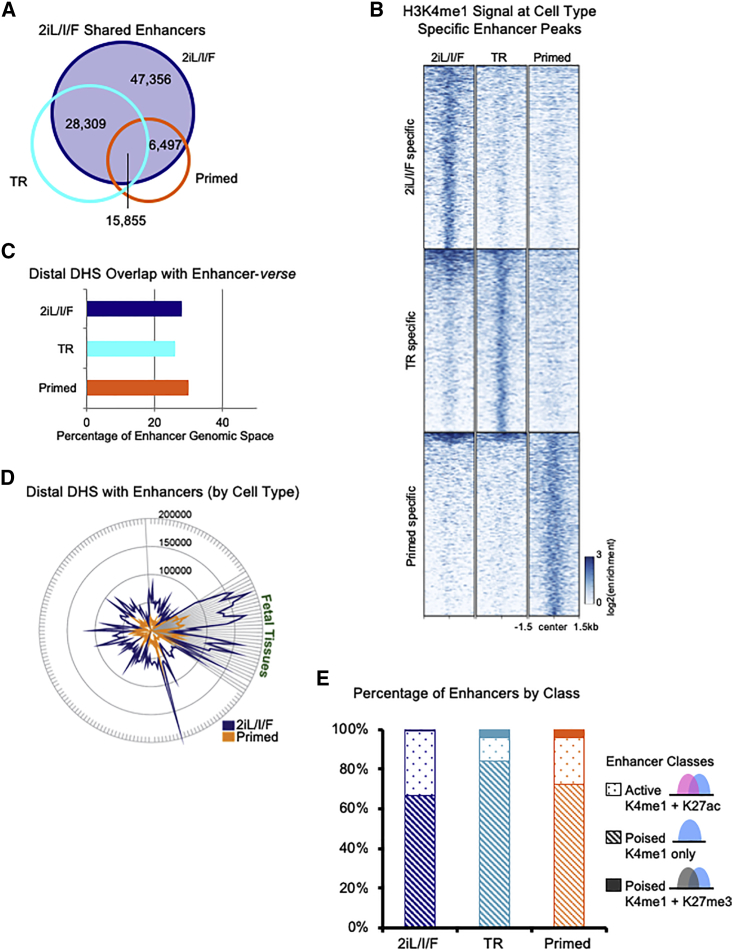
Figure 2.
2iL/I/F Enhancer Repertoire
(A) Venn diagram of 2iL/I/F (navy) enhancers overlapped with transitioning (cyan) and primed (orange) enhancers.
(B) Heatmap of H3K4me1 normalized ChIP-seq signal centered at cell type-specific enhancers in a 3-kb window.
(C) Percentage of hESC H3K4me1 genomic space, i.e., number of base pairs, occupied by ENCODE DNase hypersensitive sites (DHS) from 177 cell types.
(D) Number of ENCODE DHS from 177 cell types overlapping with 2iL/I/F and primed H3K4me1 enhancers.
(E) Percentage of active (H3K4me1 + H3K27ac) and poised (H3K4me1 only or H3K4me1 + H3K27me3) enhancer states in each cell type.