Figure 5.
3D Genome Architecture in 2iL/I/F hESCs
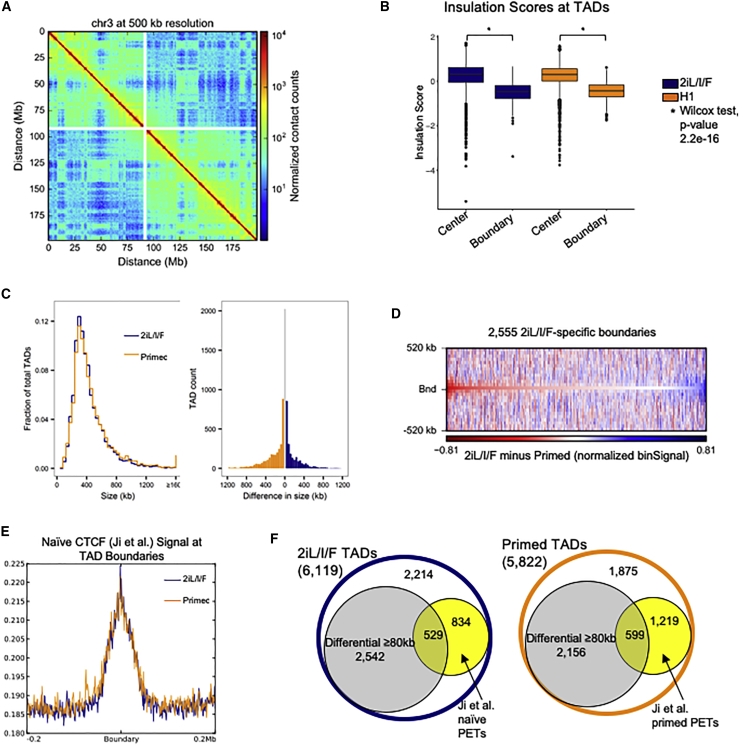
(A) Hi-C contact heatmap of chromosome 3 in 2iL/I/F cells at 500-kb resolution.
(B) Box plots of the insulation scores at all TAD centers and boundaries for both 2iL/I/F and primed cells. p values are computed using individual Wilcoxon signed-rank tests.
(C) Global size distributions of TADs within 2iL/I/F and primed cells (left panel) and size differences of overlapping TADs (40-kb bin resolution) in 2iL/I/F and primed cells (right panel).
(D) Differential heatmap of 2iL/I/F minus primed Hi-C. Negative (red) values indicate a stronger bin signal in primed cells relative to 2iL/I/F cells.
(E) Naive CTCF ChIP-seq signal from Ji et al. (2016) centered at 2iL/I/F TAD boundaries.
(F) Number of TADs or differential TADs with cohesin ChIA-PETs within 80 kb of boundary.