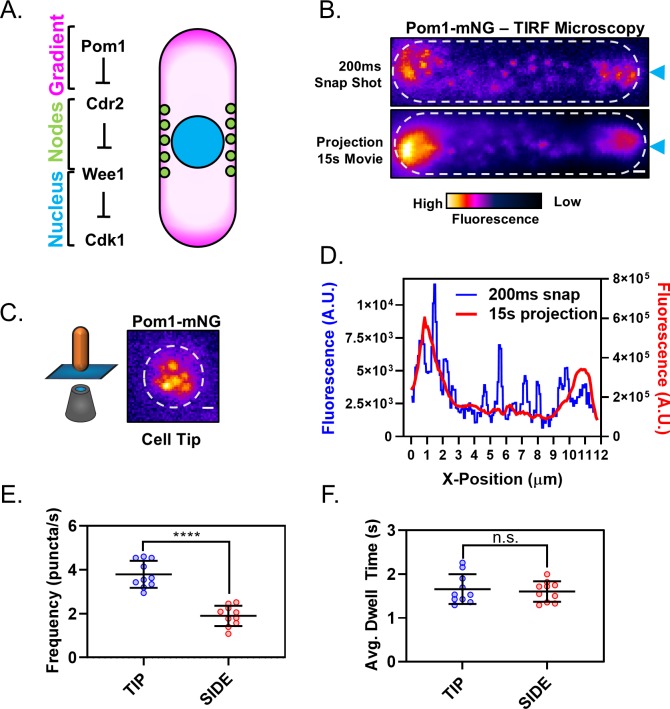
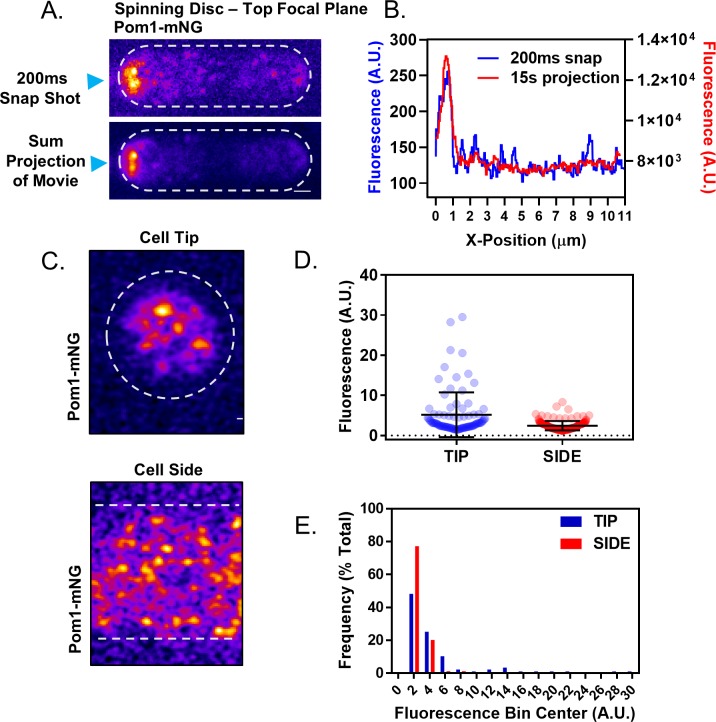
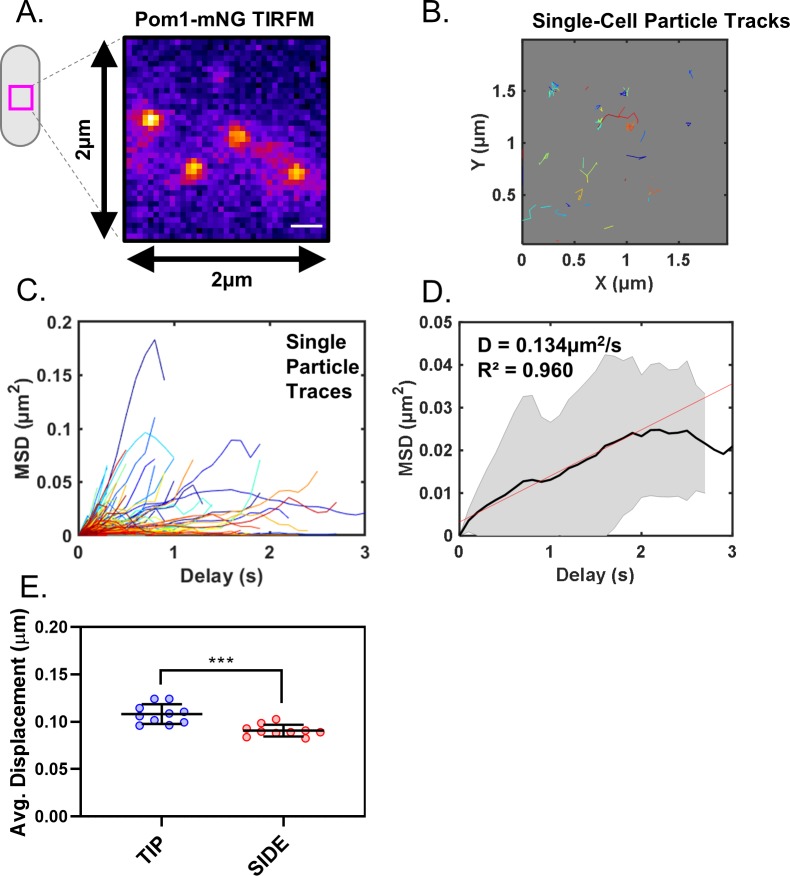
Figure 1. The Pom1 gradient is formed by time-averaging of clusters that transiently bind the cortex.
(A) Schematic of the Cdr2-Pom1-Wee1 signal transduction pathway and its coarse cellular localization. (B) Individual frame (top panel) and sum projection (bottom panel) of a high-speed TIRF microscopy movie. Movie was continuous 15 s time-lapse acquisition of 200 ms exposures. Scale bar 1 µm. Blue arrows mark position of line scans performed for data in panel (D). (C) Pom1-mNG also forms clusters at the cell tip. Image is a sum projection of three consecutive 200 ms exposures from continuous time-lapse TIRF movie of the cell tip as depicted in the cartoon diagram. Scale bar 1 µm. (D) Line scans of fluorescence intensity along the long axis of the snap shot (blue line, left Y axis) and projection images (red line, right Y axis) in panel (B). Note that time-averaging of Pom1 clusters smoothens the concentration gradient. (E) Comparison of Pom1 cluster binding frequency at the cell tip or side (****p=<0.0001, n = 10 cells, 42–170 traces/cell). (F) Comparison of cortical dwell time of individual Pom1-mNG clusters (n.s., p=0.6747, n = 10 cells, 42–170 traces/cell) at the cell tip or side. For (E–F), each data point represents a single cell mean, and line and error bars represent mean and standard deviation of all cells. Statistical significance was tested using a Student’s T-test.