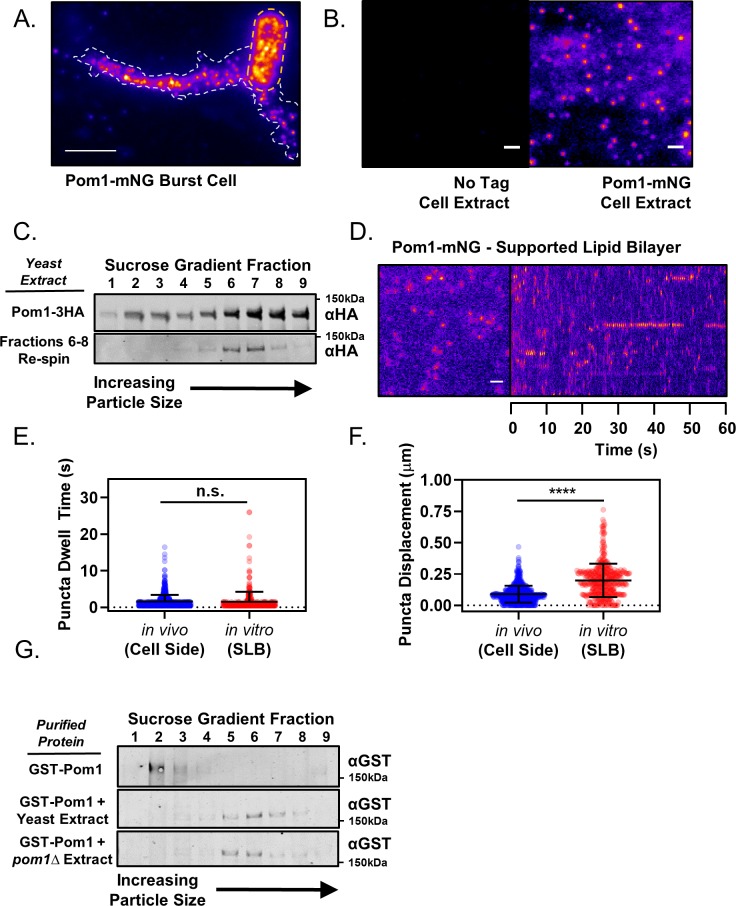
Figure 2. Pom1 clusters are stable structures that can be isolated in vitro.
(A) TIRF microscopy image of Pom1-mNG clusters in the extruded cytoplasm (white dotted line) of a lysed cell (yellow dotted line). Scale bar 5 µm. (B) TIRF microscopy images of cell extracts prepared from wild-type (no tag) or Pom1-mNG cells. Images are 50 frame sum projections of continuous 200 ms time-lapse exposures. The two images were contrasted equally. Scale bars 1 µm. (C) Cytoplasmic extracts of pom1-3HA cells were subjected to velocity sucrose gradient sedimentation, and fractions were probed against the HA tag (upper blot). Fraction one corresponds to the top of the gradient and contains smaller structures; fraction nine corresponds to bottom of the gradient and contains larger structures. Fractions 6–8 were pooled, sucrose was removed by dialysis, and then the sample was subjected to a second identical round of sucrose gradient sedimentation and western blotting of the resulting fractions (lower blot). (D) TIRF microscopy of Pom1-mNG clusters from cytoplasmic extracts on supported lipid bilayers. Scale bar 1 µm. Left panel is single time point image. Right panel is kymograph taken from a line scan of time-lapse TIRF experiment. (E) Quantification of binding duration of Pom1-mNG clusters on supported lipid bilayers imaged by TIRF microscopy as in panel (D). Values are compared to cellular measurements of Pom1 clusters on cell sides (n.s., p=0.05954, n = 713 in vivo, 421 in vitro). (F) Quantification of life-time displacement of Pom1-mNG clusters diffusing on supported lipid bilayers imaged by TIRF microscopy as in panel (D). Values are compared to cellular measurements of Pom1 clusters on cell sides (****p<0.0001, n = 713 in vivo, 421 in vitro). For (E) and (F), statistical significance was tested using a Student’s T-test. (G) Purified GST-Pom1 was subjected to sucrose gradient sedimentation and the fractions were probed against the GST tag (upper blot). Purified GST-Pom1 was also added to wild-type or pom1∆ cell extracts and incubated for 1 hr at 4 ˚C in the presence of ATP before velocity sucrose sedimentation and western blotting (bottom blots).