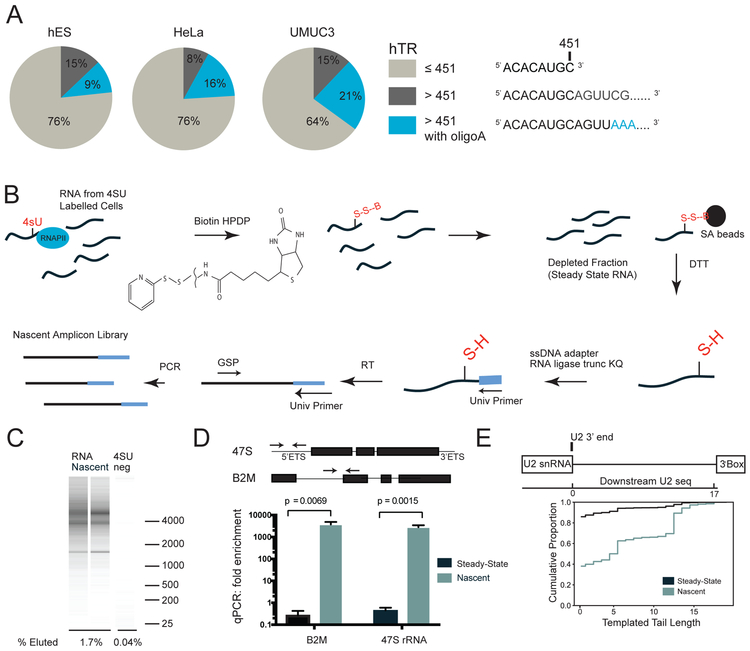
Figure 1: Identification of RNA precursors using nascent RNAend-Seq.
A. 3’ RACE-Seq of hTR in human embryonic stem (hES), HeLa, and UMUC3 cells. hTR transcripts either end at the annotated end of 451 nts or shorter (brown), are >451 nts with genomically templated tails (grey), or are >451 nts with untemplated oligo-A tails (blue). hTR Reads≥200k
B. Nascent RNAend-Seq to enrich nascent RNA tails. 4SU labeled RNA from pulsed cells is conjugated to biotin (Biotin-HPDP, B). Biotin-conjugated nascent RNA is captured with streptavidin (SA) beads and separated from steady state (SS) RNA. Nascent RNA is eluted with DTT, ligated with a 3’ ssDNA adapter, reverse transcribed (RT) and amplified with a gene-specific primer (GSP)
C. RNA bioanalyzer shows 1.7% of 4SU labeled input RNA is recovered vs. 0.04% of 4SU-unlabeled RNA.
D. qRT-PCR for RNA precursors including 47S rRNA and beta-2-microglobulin (B2M) in nascent RNA (blue) and steady state RNA (black). 47S rRNA primers span 5’ externally transcribed spacer (ETS) and B2M primers span an intron-exon junction. Data normalized to exonic GAPDH primers. Error bars represent S.E.M.
E. Sequencing of nascent U2 3’ end with the proportion of reads having a given tail length. Height of the line at templated tail length=0 indicates the proportion of reads with no tail; height of the line at templated tail length=10 indicated the proportion of reads with a tail≤10 nucleotides. Structure of the U2 snRNA transcription unit shown with annotated U2 3’ end graphed at 0 nts and 3’ box at 17 nts. Nascent RNAend-Seq results for U2 nascent (blue line) and steady state (black line) RNA.