Figure 5: Dyskerin and the Box/HACA element confer RNA end formation and regulation by PARN.
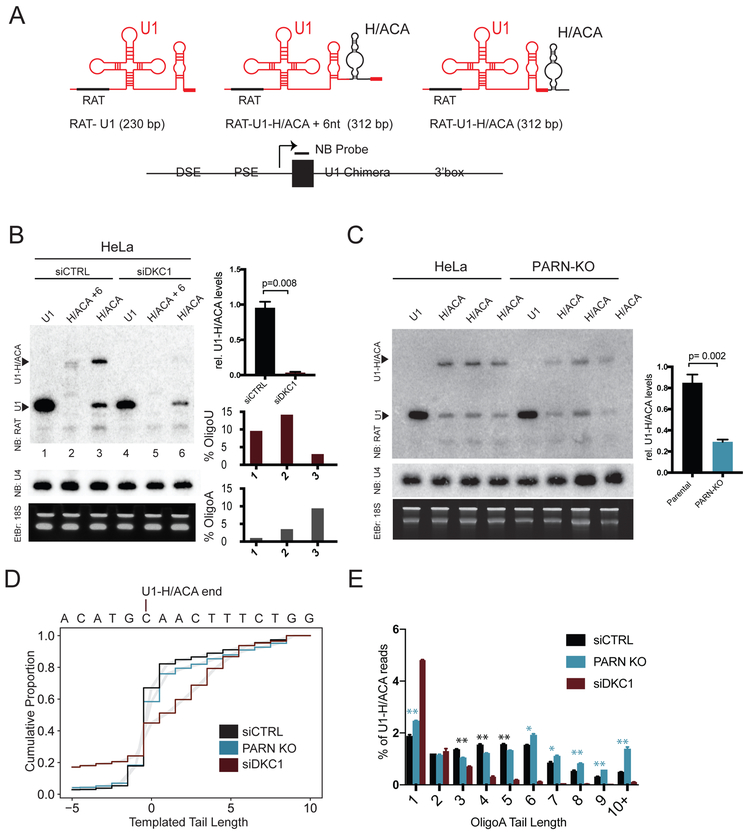
A. hTR H/ACA is grafted to the 3’ end of U1 snRNA with U1 distal sequence elements (DSE), proximal sequence elements (PSE) and the U1 3’ box. H/ACA is added at the 3’ end (U1-H/ACA) or internal (U1-H/ACA+6 nt). A RAT tag recognized by a northern blot (N.B.) probe is added in the U1 5’ region. Expected sizes indicated.
B. N.B. of chimeric RNA constructs transfected into HeLa cells. RAT-U1 indicates the band formed by termination at the U1 3’ end. U1-H/ACA indicates the band formed by termination at the chimeric U1-H/ACA end. Per lane, % of chimeric reads with untemplated A or U nucleotides (% Oligo-A, % Oligo-U) from total reads>80k. Relative U1-H/ACA levels are normalized to U4 and quantified from 2 biologic replicates. Error bars represent S.E.M.
C. N.B. of chimeric RNA constructs transiently transfected into HeLa or PARN-KO cells. Relative U1-H/ACA levels are normalized to U4 and quantified from 4 biologic replicates.
D. 3’ RACE-Seq of RAT-U1-H/ACA in cells treated with siCTRL (black), treated with siDKC1 (red), or PARN-KO cells with siCTRL (blue), with total reads>80k. The 3’ downstream sequence of the U1-H/ACA chimera is indicated on the x-axis. Gray shading is 95% CI derived from 2 sequencing replicates.
E. Percent of Oligo-A tails with the indicated lengths. Total Oligo-A reads>2.5k. Error bars represent S.E.M. Blue asterisks indicate oligoA tails with significantly more reads in PARN-KO cell lines (**p<0.01, * p<0.05). Black asterisks indicate oligoA tails with significantly more reads in parental cell lines.