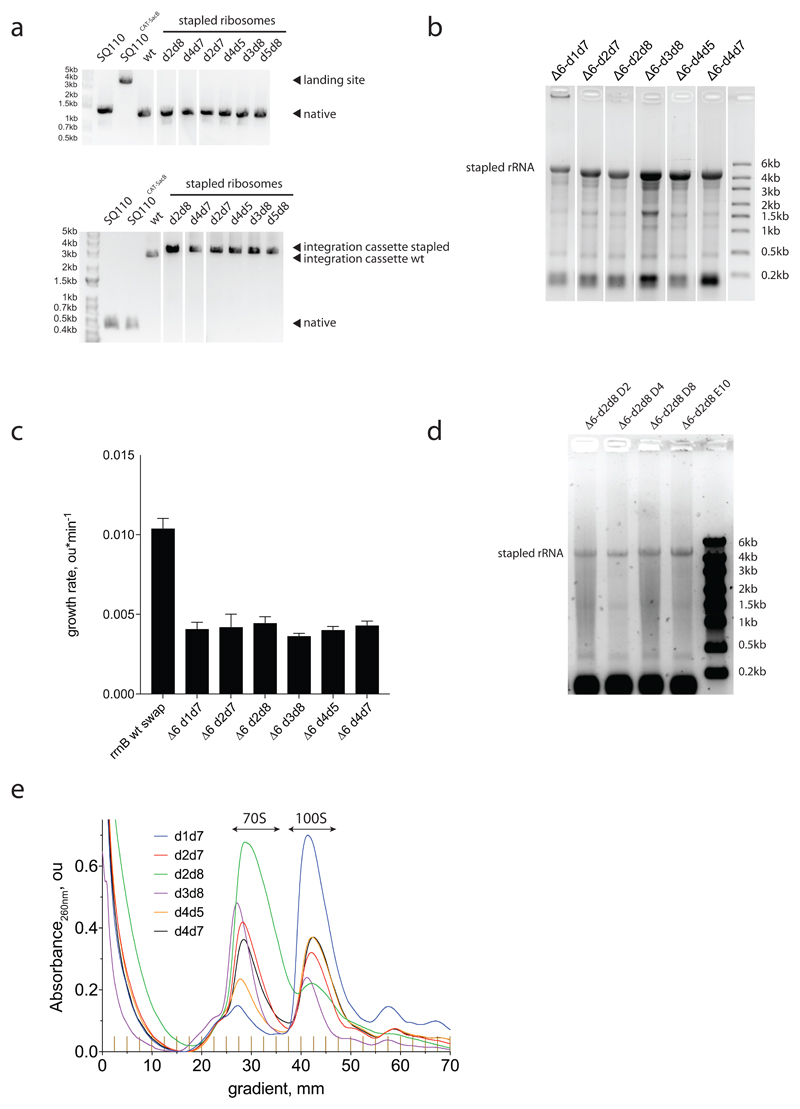
Extended Data Figure 7. Genomically encoded stapled ribosomes.
a, Colony PCR products reflect the genomic exchanges seen in E. coli strains with a genomically encoded, stapled ribosome rRNA operon integrated by ribo-REXER. Top, direct amplification of the genomic region upstream of the rrnE locus. Integration of the landing site (SacB/CAT) to create the strain SQ110CAT-SacB increases the length of the region upstream of rrnE from 1.3 kb to 3.5 kb. After ribo-REXER, this cassette is lost again, leading to a 1.3-kb band. Bottom, amplification from nucleotide 2,630 of 23S to the rrnE terminator region shows integration of the PheS*/HygR cassette after integration of the wild-type rrnB operon, indicated by ‘integration cassette wt’. Integration of a stapled-ribosome cassette increases the length of the PCR product further, as it includes the 16S 3′ and internal transcribed spacer (ITS) regions, leading to the bands indicated by ‘integration cassette stapled’. The experiment was performed once. b, Denaturing RNA gel electrophoresis reveals the expression of rRNA (around 4,500 nucleotides) from intact stapled ribosomes as the predominant RNA species in all strains. The experiment was performed once. c, Growth rates of strains after successful ribo-REXER. WT describes the integration of a non-stapled rrnB operon. For statistics, see Methods. d, Denaturing RNA gel electrophoresis shows the expression of rRNA from intact stapled ribosomes (around 4,500 nucleotides) as the predominant RNA species in all evolved Δ6 d2d8 strains. The experiment was performed once. e, Sucrose gradient analyses of stapled ribosomes isolated from cells under ribosome-associating conditions (10 mM MgCl2); the experiments were repeated three times with similar results. For source data regarding gels, see Supplementary Fig. 1.