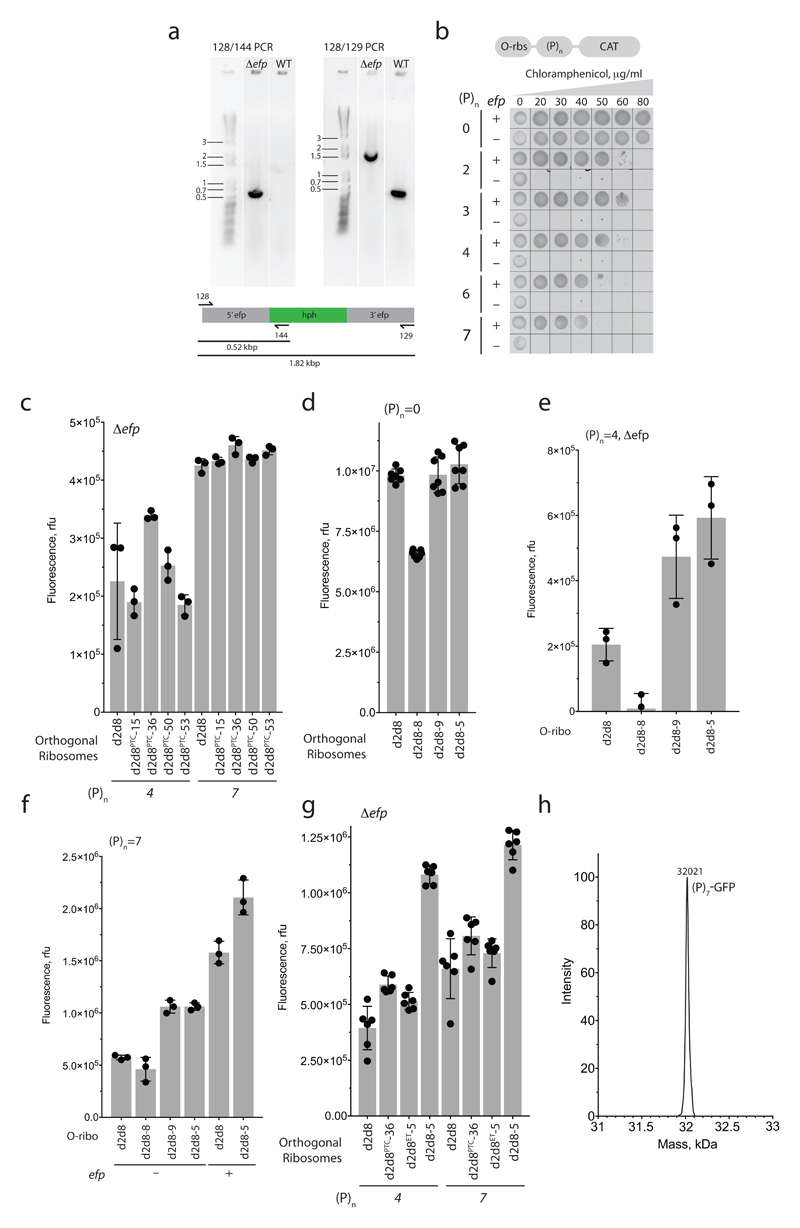
Extended Data Figure 9. Selection of O-ribosome variants competent in translation of polyproline sequences.
a, PCR characterization of an E. coli TOP10 Δefp strain. PCR was carried out, using the indicated primer pairs (128/144 or 128/129), on genomic DNA from a TOP10 strain with intact efp, or a strain in which the efp locus was disrupted by a hygromycin-B-resistance gene (hph). For source data regarding gels, see Supplementary Fig. 1. The experiment was performed twice. b, Cell-growth assay, showing the translation activity of O-d2d8 on O-(P)n-CAT reporters in wild-type (+) or Δefp (−) E. coli TOP10 cells and with varying concentrations of chloramphenicol. The experiment was performed once. c, Activity of the PTC-library hits and the parental O-d2d8 ribosome on p15A-O-(P)4-GFP and p15A-O-(P)7-GFP reporters in E. coli TOP10 Δefp cells. n = 3 biological replicates; error bars indicate ± s.d. d, Translation activity of exit-tunnel library hits in the absence of proline-rich sequences. p15A-O-GFP was used as a reporter. n = 7 biological replicates; error bars indicate ± s.d. e, GFP fluorescence resulting from translation of O-(P)4-GFP in E. coli TOP10 Δefp cells. n = 3 biological replicates; error bars indicate ± s.d. f, As for e, except that the activity of the evolved mutants was tested on an O-(P)7-GFP reporter in E. coli with and without efp. n = 3 biological replicates; error bars represent ± s.d. g, Mutations in both the PTC and the exit tunnel are required to confer on O-d2d8 the ability to translate O-(P)4-GFP and O-(P)7-GFP in TOP10 Δefp cells. O-d2d8PTC-36 and O-d2d8ET-5 contain selected mutations in the PTC and the exit tunnel respectively; O-d2d8-5 contains mutations in both. See Supplementary Data 12 for details on mutations. n = 6 biological replicates; error bars represent ± s.d. h, Electrospray ionization spectra of O-(P)7-GFP synthesized by O-d2d8-5 in Δefp E. coli. This experiment was performed once.