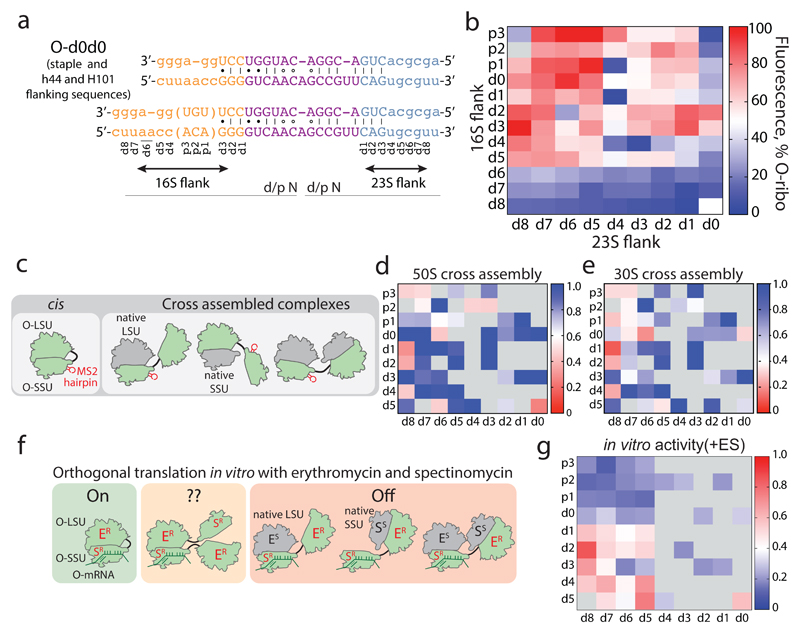
Figure 2. Maximizing activity and minimizing cross-assembly in engineered O-stapled ribosomes via systematic variation of the intersubunit linker.
a, Top two rows, the intersubunit staple sequence (uppercase letters)—composed of a hinge (purple) and native helical flanking residues (yellow and blue)—used in O-d0d0. rRNA-derived sequences are in lowercase letters. Bottom, variants are denoted by the number (N) of base pairs that have been deleted (d) or inserted (p, for ‘plus’) from the 16S side, followed by the number of base pairs deleted from the 23S side. b, Heat map showing O-stapled ribosome activity (as analysed by the synthesis of GFP) in vivo. The resulting GFP fluorescence is shown as a percentage of that produced from an orthogonal ribosome with non-linked subunits. Data are thresholded at 100. See Methods for statistics and Extended Data Fig. 1b for full data. c, Potential complexes between small subunits (SSUs) and large subunits (LSUs) following affinity purification of O-stapled ribosomes (green) with an MS2 stem loop. Native subunits are in grey. d, e Heat map showing 50S (LSU; d) and 30S (SSU; e) cross-assembly coefficients. Variants in grey were not tested. The heat map is thresholded at 1. Data are means of n=2 biological replicates. See Extended Data Fig. 3a,b. f, In vitro translation of the orthogonal message in the presence of the antibiotics erythromycin and spectinomycin (which selectively inhibit native subunits) is denoted ‘on’, ‘off ’ or unknown (‘??’) for each complex upon selective inhibition of native subunits. ER and SR denote erythromycin and spectinomycin resistance; ES and SS denote erythromycin and spectinomycin sensitivity g, Heat map showing the efficiency of translating T7-O-GFP (a construct containing a T7 promoter upstream of an orthogonal ribosome-binding site and an sfGFP gene) in S30 extracts of E. coli; the extracts contained the indicated O-stapled ribosome and native ribosomes. We added 10 μM spectinomycin and 50 μM erythromycin (ES) to the extract to inhibit native subunits. The heat map shows the mean fluorescence. For values of n and errors, see Methods and Extended Data Fig. 6.