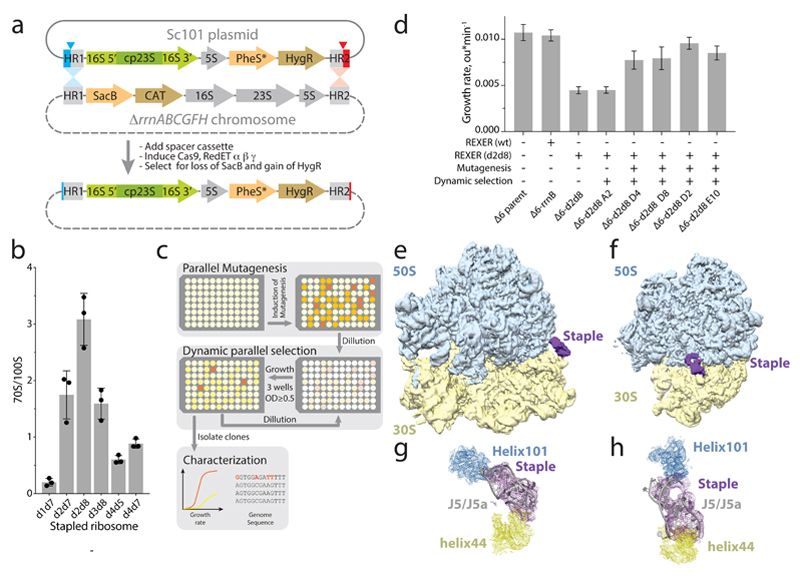
Figure 3. Genomically encoding stapled ribosomes as the sole cellular ribosomes, and subsequent strain evolution, generates fast-growing E.coli for d2d8 purification and structure determination.
a, Schematic showing how ribo-REXER is used for the genomic replacement of ribosomal RNA operons, applied here to generate Δ6d2d8. The SC101 plasmid contains the rRNA for the stapled ribosome; this rRNA is used to replace the single rRNA in the chromosome of a ΔrrnABCGFH strain of E. coli. SacB, Bacillus subtilus levansucrase gene; CAT, chloramphenicol acetyltransferase gene; cp, circularly permuted; HR, homologous regions; HygR, hygromycin-resistance gene; PheS*, T251A A294G mutant of E. coli phenylalanyl-tRNA synthetase. Red and blue arrows indicate sites of Cas9-mediated cleavage. b, Ratio of monosomes (70S) to ribosome dimers (100S) from sucrose gradient analyses of different stapled ribosomes isolated from cells under associating conditions. Shown are individual data points (black dots), means (grey bars) and standard deviations from three biological replicates. c, Evolution of Δ6 d2d8 by automated parallel evolution. Clonal cultures are shown in pale yellow; mutagenized populations in each well are represented by distinct shades. Dynamic parallel selection was repeated for ten cycles before clone isolation and genome characterization. OD indicates the optical density at 600 nm. In the dynamic parallel selection, the dilution of cells was triggered once the OD600 of three wells was greater than or equal to 0.5. d, Characterization of the specific growth rates of Δ6 d2d8 (resulting from ribo-REXER), as well as passaged passaged (A2) and evolved (D4, D8, D2, E10) strains. rrnB is an rRNA operon For statistics, see Methods. e, Electron-density map for E.coli d2d8 70S ribosome. f, A rotated view of the electron-density map shown in e, g. A close up of the density (purple mesh) connecting Helix 101 and helix 44. The J5-J5a RNA (Protein Data Bank (PDB) accession code 1GID; grey) is docked into the density for the staple. The orientation shown is comparable to that in e. h, Similar to g, but shown in a comparable orientation to f.