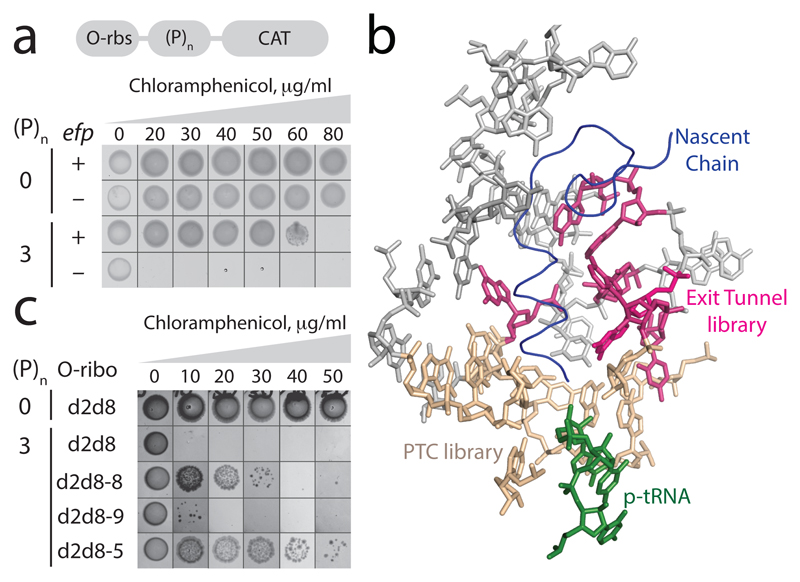
Figure 4. Discovering O-d2d8 variants with the intrinsic ability to translate polyproline sequences.
a, Top, the O-rbs–(P)n–CAT gene, in which an orthogonal ribosome-binding site (O-rbs) directs the translation of proline (P) codons (n=0 or 3) followed by the chloramphenicol acetyltransferase (CAT) gene. When the number of proline codons is 0, the O-d2d8 ribosome can translate the CAT gene even in the absence of efp (encoding EF-P), and thus confers resistance to chloramphenicol. In the absence of efp, the ribosome cannot translate through the polyproline (n = 3) sequence and thus cannot translate the CAT gene; hence the strain is not resistant to chloramphenicol and does not grow. The experiment was performed twice with similar results. b, The region of the ribosome that we targeted for mutation (PDB accession code 5NWY) in order to facilitate translation of polyproline sequences. Beige, nucleotides (C2063, G2447, A2450, A2451, C2452, U2506, G2583, U2584, U2585 and A2602) of the 23S rRNA in the peptidyltransferase centre (PTC) that were randomized. Pink, nucleotides (2058A, 2059A, 2061G, 2062A, 2501C, 2503G and 2505G) in the exit tunnel that were randomized. c, Chloramphenicol resistance provided by evolved O-d2d8 variants in the absence of EF-P. The experiment was performed twice with similar results. Note that cells grow more slowly in strains lacking EF-P.