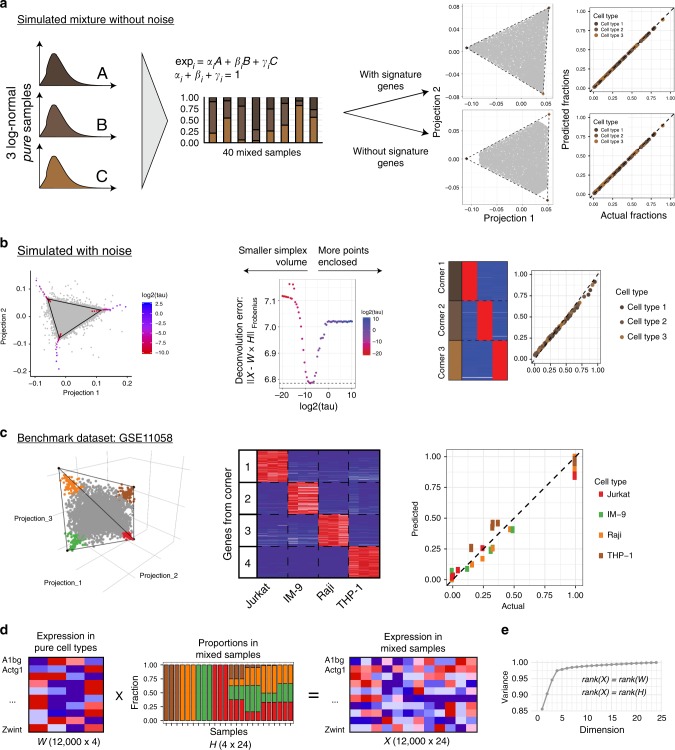
Fig. 4.
Geometric algorithms for simplex identification accurately solve the complete deconvolution problem in benchmark and simulations datasets. a Application of the algorithm to a simulated mixture. Left: dataset design—mixture of three pure samples with log-normally distributed gene expression. Right: Identification of simplex corners with and without signature genes: SISAL robustly identifies corners in both cases. b Dependence of deconvolution accuracy on tolerance level (tau) for simulated data with noise (left and middle). Deconvolution results for best tau (right): heatmap of corner genes expression in pure cell types and comparison of actual versus estimated cell fractions. c Application of algorithm to benchmark dataset GSE11058: identified simplex has a form of tetrahedron (4-simplex), heatmap shows cell-specific expression of corner genes and dot plot shows concordance between actual and estimated fractions of cell types. d Schematic for linear model for dataset GSE11058. e Variance explained by each SVD component for benchmark dataset GSE11058. Expression levels in heatmaps in panel b and c are from 0 (blue) to gene max (red)