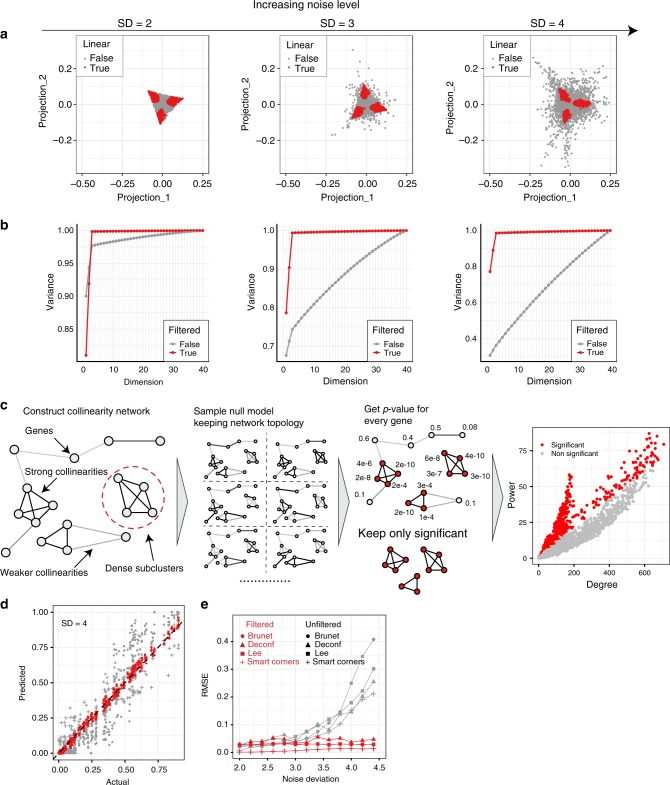
Fig. 5.
Mutual linearity filtering improves deconvolution methods and allows cell type number identification in noisy cases. For panels a and b noise level is increasing from left to right. a Two-dimensional projection of simulation with noise. Points in red are selected by filtering procedure. b Variance explained by each SVD component for both unfiltered (gray) and filtered (red) datasets. c Schematic for filtering approach. Collinearity network is built by calculating all the pairwise linearity coefficients and spearman correlations. When the network is built, weights of the edges are randomly shuffled to test each gene to have more total weight than at random. We select only those genes with a significance level < 0.01. On the right is illustration of which genes are selected, where power denotes the sum of the weights of the edges. d For very noisy dataset linearity filtering improves performance of all complete deconvolution methods. e For different noise levels RMSE of predicted and actual proportions is calculated