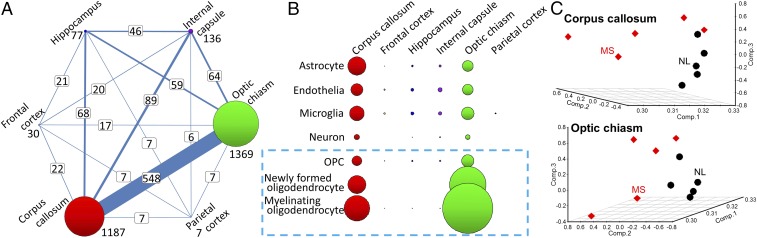
Fig. 1.
Gene-expression changes in MS. High-throughput RNA sequencing was performed for six different brain regions from patients with MS (n = 5) and healthy controls (n = 5). (A) Transcriptome changes in the CNS of MS are region-specific. The numbers of differentially expressed genes in MS vs. controls are displayed as the sizes of circles, with differentially expressed gene numbers listed next to them. The numbers of shared differentially expressed genes between two different CNS tissues are shown in white boxes. The thickness of lines connecting two tissues reflects the level of sharing of differentially expressed genes. The transcriptome changes by disease were striking in corpus callosum and optic chiasm, and nearly half of differentially expressed genes were shared between these two tissues. (B) By using the top 500 CNS cell markers for seven different cell types (astrocyte, endothelia, microglia, neuron, OPC, newly formed oligodendrocyte, and myelinating oligodendrocyte) from the RNA-sequencing transcriptome database (11), the genes enriched in each cell type were selected, and the differentially expressed gene numbers (false discovery rate < 0.1) are visualized as the sizes of circles. Oligodendrocyte markers in corpus callosum and optic chiasm showed significant transcriptome changes by disease. (C) Principal component analysis of MS and control (NL) samples from corpus callosum and optic chiasm. Separation of MS and normal samples was observed in corpus callosum (Top) and optic chiasm (Bottom). MS group, five women with progressive MS, mean age, 57.6 y; control, five women, mean age, 56.2 y. Based on these results in MS, Olig1-RiboTag mice were used to define the transcriptome in OLCs (light blue dashed box in B) in corpus callosum.