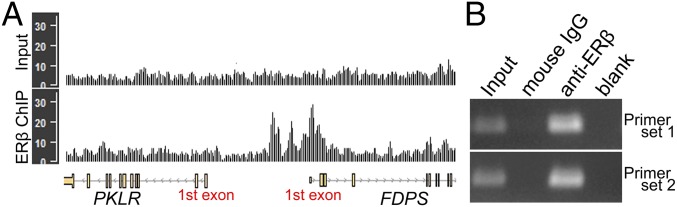
Fig. 10.
ERβ-binding profiles for human FDPS and ChIP assay for mouse Fdps gene. (A) ChIP-seq data for doxycycline-inducible ERβ-expressing human MDA-MB-231 cells treated with estradiol was obtained from the GEO database (accession no. GSE108981). Peaks in ERβ ChIP profiles indicate ERβ binding on the region. FDPS showed peaks around the first exons in ERβ ChIP profile, whereas the negative control input profile (DNAs from chromatin before ChIP) did not show the peak. As an additional negative control, the PKLR gene, near the FDPS gene, did not have a peak. (B) ERβ binds to the putative ERE of mouse Fdps gene. ChIP using normal mouse IgG and anti-ERβ was performed for the chromatin isolated from mouse N20.1 oligodendrocyte cell line (21) treated with ERβ ligand (DPN). Two sets of primers were designed across the region containing putative ERE of Fdps gene (51).