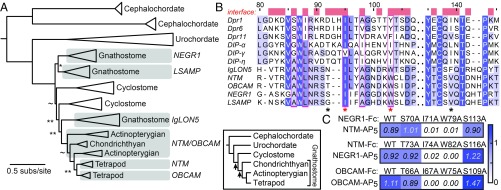
Fig. 2.
IgLONs are the vertebrate Wirins. (A) The ML phylogeny of the IgLON subfamily. aLR statistics are shown as branch supports: **<9.2 (= 2 ln100), *< 4.6 (= 2 ln10), ∼<2.2 (= 2 ln3). Unmarked branches have aLR statistics >9.2. Inset shows the established chordate phylogeny, with arrows marking two genome duplications. (B) Sequence alignment of the IG1 domains of mouse IgLONs, three Dprs, and three DIPs. Amino acids at the Dpr-DIP interface, defined by a 4-Å cutoff from the binding partner, are labeled as red blocks above the alignment. Amino acids mutated in C are labeled with an asterisk. The hydrophobic core residues of the Dpr-DIP interface observed by Cheng et al. (11) is indicated by magenta columns. Sequence numbers above the alignment are for Dpr1. (C) Mutations at the predicted interfaces of the OBCAM-OBCAM and NTM-NEGR1 complexes affect binding as tested using ECIA. To effectively compare WT to mutants, protein concentrations within each mutant series were normalized. Reported absorbance values are after subtraction of negative controls at 0.05 (±0.01) absorbance units.