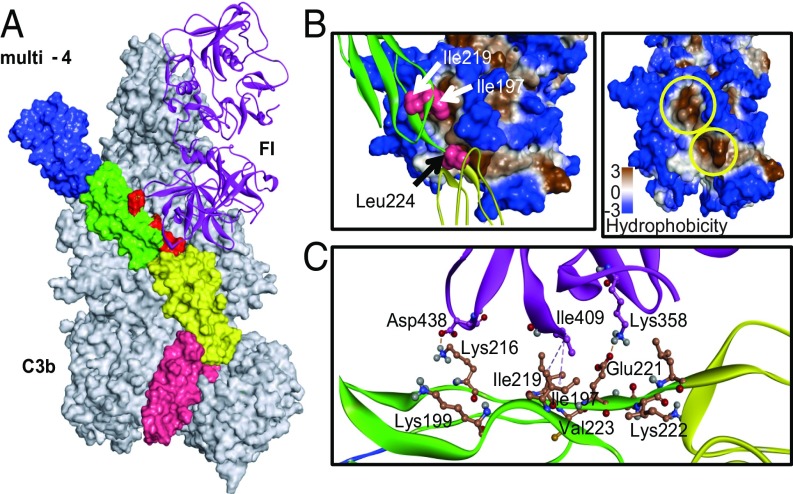
Fig. 5.
Mapping of factor I interaction sites in the C3b-multi-4 mutant-FI trimolecular complex. (A) Model of the C3b-multi-4 mutant-FI trimolecular complex. The D2D3M3M4 chimera having all of the CFA gain-of-function residues (multi-4 mutant) was superimposed with the coordinates of FH in the C3b-FH-FI crystal structure (Protein Data Bank ID code 5O35). The model was then subjected to MD simulations for 50 ns and analyzed for the interactions of gain-of-function residues with FI. Each of the four domains of the multi-4 mutant (shown as surface) was labeled with different colors (CCP1-blue, CCP2-green, CCP3- yellow, CCP4-pink). C3b (shown as surface) and FI (ribbon representation) are in gray and magenta, respectively. Gain-of-function residues are in red. (B) Zoomed view of the interactions shown by the gain-of-function residues with FI. (Left) I197 and I219 sit in the upper hydrophobic pocket formed by the residues W393, P402, L404, I407, V408, I409, and Y411, while L224 sits in a lower hydrophobic pocket formed by residues I357, G362, I363, A360, V396, V397, W399, and I400. (Right) The hydrophobic pockets shown on the Left are shown again and marked with yellow circles. (C) Charge and hydrophobic interactions of gain-of-function residues with FI.