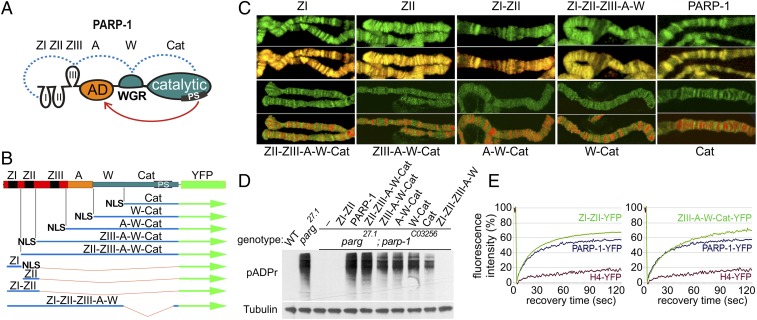
Fig. 1.
N- and C-terminal PARP-1 domains contribute to PARP-1 protein localization genome-wide in vivo. (A) Domains of PARP-1: The N-terminal DNA-binding domain (DBD) containing Zn fingers: ZI, ZII, ZIII (ZF domains 1, 2, and 3), the automodification domain (A), the only domain of PARP-1 known to accept pADPr, the WGR domain (W), and the C-terminal catalytic domain (C). The PARP signature (PS) is an evolutionarily conserved PARP-1 catalytic site in the Cat domain. The dotted blue line indicates known interactions between the domains induced by interactions with damaged DNA (21). The red arrow indicates automodification of PARP-1. (B) Structure of recombinant-transgenic PARP-1 constructs for in vivo experiments. (C) Localization of deletional recombinant isoforms of PARP-1 in salivary gland polytene chromosomes. Green is the fluorescence of proteins fused to YFP, red is DNA. All isoforms of PARP-1 carrying ZI and ZII demonstrate colocalization with DNA resulting in the yellow color in the overlay. All isoforms without ZI and ZII localized in active open chromatin only, resulting in the separation of red and green in the overlay. (D) PARP-1 deletional isoform activity assay in vivo. PARP-1 deletional isoforms were expressed in the parg27.1; parp-1C03256 mutant flies. All isoforms containing the Cat domain restored pADPr accumulation. (E) Both DNA- and C-terminal domains contributed to PARP-1 protein dynamic binding to chromatin in vivo. Comparative analysis of fluorescent recovery after photobleaching (FRAP) assay for recombinant protein is shown, including ZI-II-YFP, ZIII-A-W-Cat-YFP, full-length PARP-1-YFP, and H4-YFP. Data for the FRAP experiment show the average based on 10 replicates.