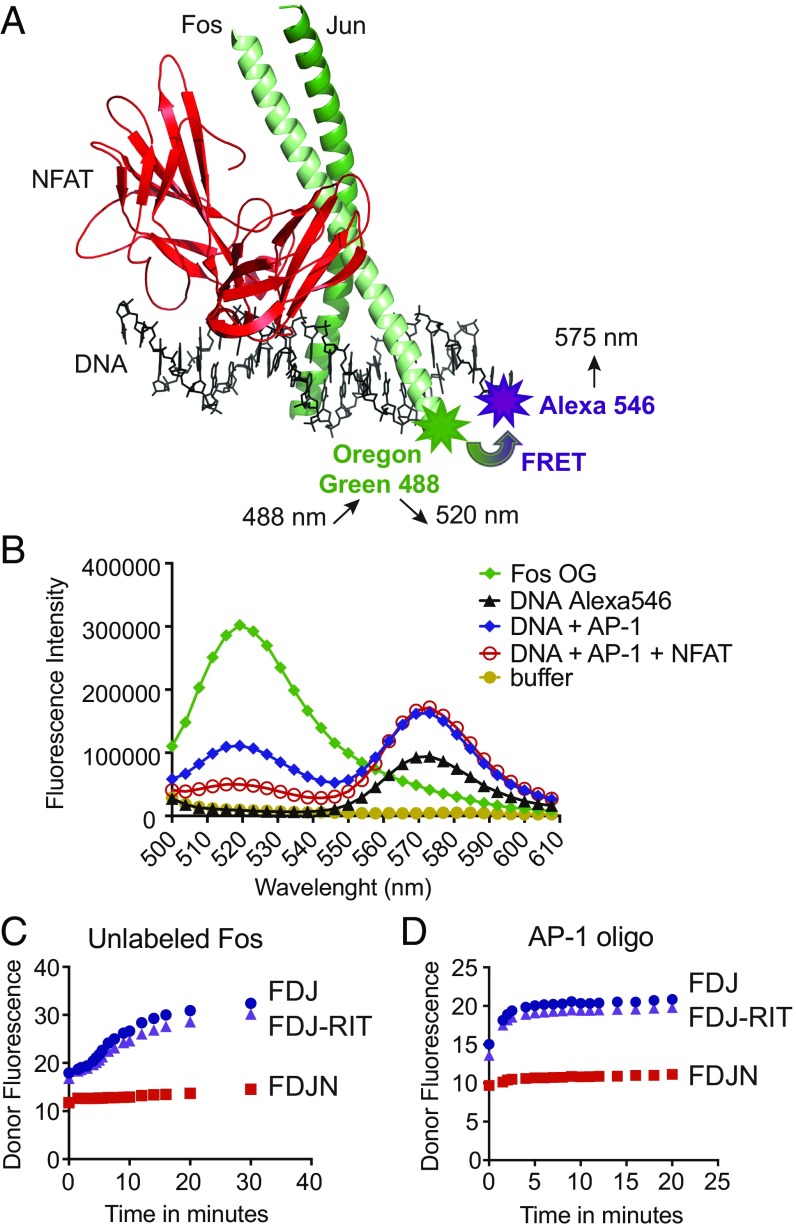
Fig. 1.
Design of the FRET assay. (A) Structure of the NFAT:AP-1 complex bound to the ARRE-2 element of the murine IL-2 promoter (PDB ID code 1A02). The DBD of NFAT (red) is in contact with the AP-1 dimer of Fos (light green) and Jun (dark green). The NFAT site in the ARRE-2 oligonucleotide is a consensus binding site for NFAT, whereas the AP-1 site is a nonconsensus site that differs appreciably from the consensus AP-1 site TGAC/GTCA. Modified from figure 2 in ref. 35. Fos was labeled with the donor fluorophore Oregon Green 488 maleimide. The 3′ end of the sense strand of the ARRE-2 DNA oligonucleotide was labeled with the acceptor fluorophore Alexa-546. (B) Fluorescence emission scan of the donor Fos-OG alone (emission peak at 520 nm, green curve), the acceptor DNA-Alexa 546 alone (emission peak at 575 nm, black curve), and the indicated protein–DNA complexes (blue and red curves). The assay was excited at 488 nm; concentrations used were 20 nM Alexa-546–labeled ARRE-2–DNA, 20 nM Fos-OG, 20 nM Jun, and 40 nM NFAT1 DBD. (C and D) Dissociation kinetics of the loosely bound Fos:Jun:DNA (FJD) and Fos:Jun:DNA:NFAT-RIT (FJD-RIT) complexes versus the cooperatively bound Fos:Jun:DNA:NFAT (FJDN) complex upon the addition of 200 nM unlabeled Fos (C) or 20 nM consensus AP-1 oligonucleotide (D). The initial complexes had been assembled from 20 nM Fos-OG, 20 nM ARRE-2–Alexa546 DNA, and 20 nM Jun, with the inclusion of 40 nM wild-type NFAT DBD or 40 nM NFAT-RIT DBD where indicated. Donor fluorescence is plotted. The assay was read using a Synergy 2 (Biotek) plate reader with 485/20-nm and 528/20-nm filters for excitation and emission, respectively.