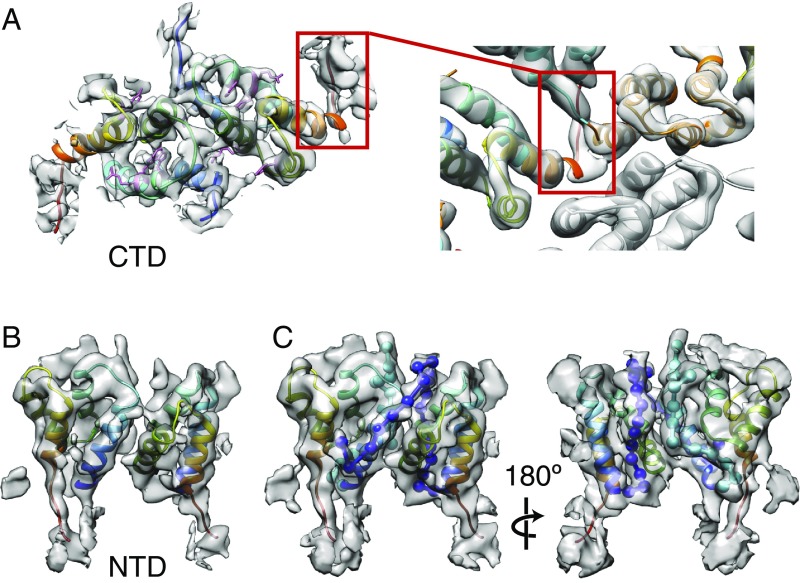
Fig. 4.
Ty3 CA-NTD and CA-CTD at higher resolution. (A, Left) Structure of a Ty3 CA-CTD dimer at 4.9 Å generated by alignment and averaging of nine non–symmetry-related copies of the Ty3 CA-CTD. The homology models of the Ty3 CA domains have been flexibly fitted into the refined EM maps. Protein models are colored from blue (N terminus) to red (C terminus). The bulky side chains of F134, R135, W138, R157, and Y164 amino acids in the CA-CTD are colored pink. (A, Right) Close-up view of the very C-terminal part of the CA-CTD from the complete Ty3 particle map, which creates one of the contacts between two neighboring CA monomers in the particle lattice. (B) Structure of a CA-NTD dimer at 5.5 Å generated by alignment and averaging of four non–symmetry-related copies of the Ty3 CA-NTD dimer, colored as in A. (C) As in B, but shown at a lower isosurface level to illustrate the unoccupied densities in the refined EM map of the CA-NTD dimer. These represent the very N-terminal parts of the Ty3 CA-NTD. The N-terminal part of the CA-NTD is shown as a string of beads in cyan (conformation A) and blue (conformation B). In both conformations, the N-terminal part of the protein first runs outward along the inner interface between the CA-NTD helix 1 and helix 2 (C, Right). In conformation B, it then continues over the top of the CA-NTD and down the outer side of the threefold position (C, Left).